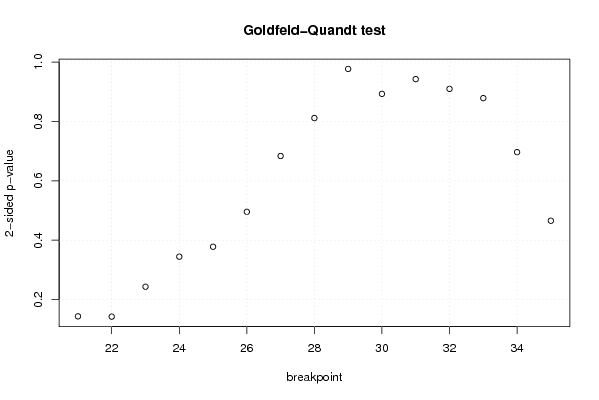
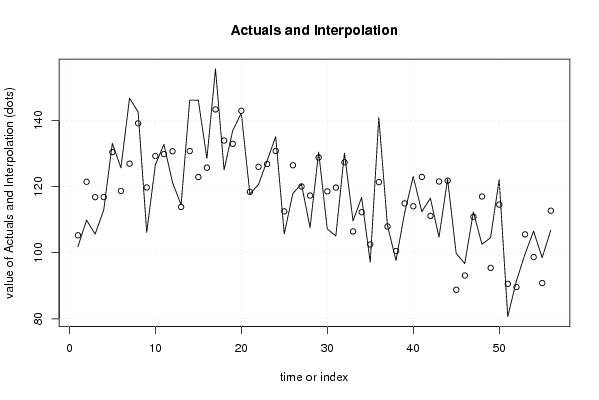
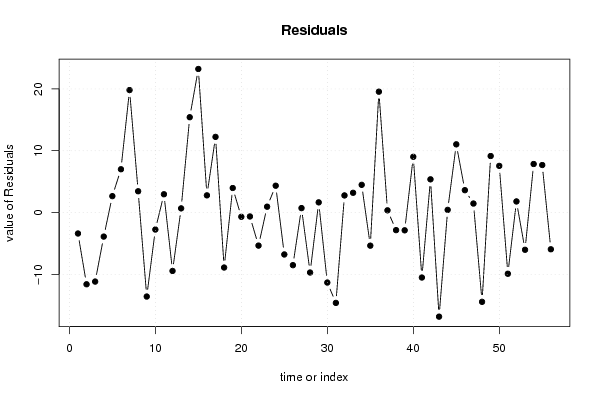
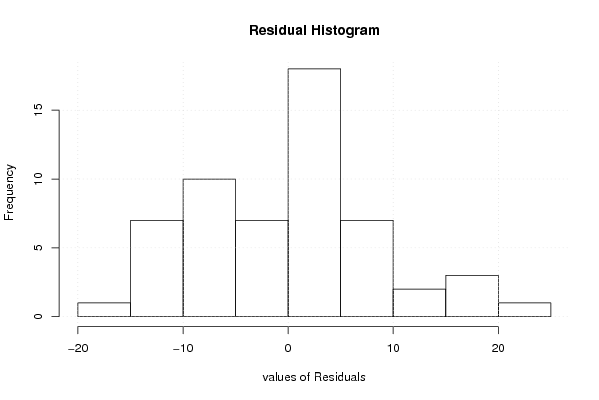
101,8612953 1 118,1540031 105,5073942 95,84395716 100
109,8419174 1 101,8612953 118,1540031 105,5073942 95,84395716
105,6348802 1 109,8419174 101,8612953 118,1540031 105,5073942
112,927078 1 105,6348802 109,8419174 101,8612953 118,1540031
133,0698623 1 112,927078 105,6348802 109,8419174 101,8612953
125,6756757 1 133,0698623 112,927078 105,6348802 109,8419174
146,736359 1 125,6756757 133,0698623 112,927078 105,6348802
142,5803162 1 146,736359 125,6756757 133,0698623 112,927078
106,1448241 1 142,5803162 146,736359 125,6756757 133,0698623
126,5170831 1 106,1448241 142,5803162 146,736359 125,6756757
132,7893932 1 126,5170831 106,1448241 142,5803162 146,736359
121,2391637 1 132,7893932 126,5170831 106,1448241 142,5803162
114,5079041 1 121,2391637 132,7893932 126,5170831 106,1448241
146,1499235 1 114,5079041 121,2391637 132,7893932 126,5170831
146,1244263 1 146,1499235 114,5079041 121,2391637 132,7893932
128,5058644 1 146,1244263 146,1499235 114,5079041 121,2391637
155,5838858 1 128,5058644 146,1244263 146,1499235 114,5079041
125,0382458 1 155,5838858 128,5058644 146,1244263 146,1499235
136,8944416 1 125,0382458 155,5838858 128,5058644 146,1244263
142,2233554 1 136,8944416 125,0382458 155,5838858 128,5058644
117,7715451 1 142,2233554 136,8944416 125,0382458 155,5838858
120,627231 1 117,7715451 142,2233554 136,8944416 125,0382458
127,7664457 1 120,627231 117,7715451 142,2233554 136,8944416
135,1096379 1 127,7664457 120,627231 117,7715451 142,2233554
105,7113717 1 135,1096379 127,7664457 120,627231 117,7715451
117,9245283 1 105,7113717 135,1096379 127,7664457 120,627231
120,754717 1 117,9245283 105,7113717 135,1096379 127,7664457
107,572667 1 120,754717 117,9245283 105,7113717 135,1096379
130,4436512 1 107,572667 120,754717 117,9245283 105,7113717
107,2157063 1 130,4436512 107,572667 120,754717 117,9245283
105,0739419 1 107,2157063 130,4436512 107,572667 120,754717
130,1121877 1 105,0739419 107,2157063 130,4436512 107,572667
109,6379398 1 130,1121877 105,0739419 107,2157063 130,4436512
116,7261601 1 109,6379398 130,1121877 105,0739419 107,2157063
97,11881693 0 116,7261601 109,6379398 130,1121877 105,0739419
140,8975013 1 97,11881693 116,7261601 109,6379398 130,1121877
108,2865885 1 140,8975013 97,11881693 116,7261601 109,6379398
97,65425803 0 108,2865885 140,8975013 97,11881693 116,7261601
112,0346762 1 97,65425803 108,2865885 140,8975013 97,11881693
123,0494646 1 112,0346762 97,65425803 108,2865885 140,8975013
112,4171341 1 123,0494646 112,0346762 97,65425803 108,2865885
116,4966854 1 112,4171341 123,0494646 112,0346762 97,65425803
104,6914839 1 116,4966854 112,4171341 123,0494646 112,0346762
122,2335543 1 104,6914839 116,4966854 112,4171341 123,0494646
99,79602244 0 122,2335543 104,6914839 116,4966854 112,4171341
96,71086181 0 99,79602244 122,2335543 104,6914839 116,4966854
112,3151453 1 96,71086181 99,79602244 122,2335543 104,6914839
102,5497195 1 112,3151453 96,71086181 99,79602244 122,2335543
104,5385008 1 102,5497195 112,3151453 96,71086181 99,79602244
122,0805711 1 104,5385008 102,5497195 112,3151453 96,71086181
80,64762876 0 122,0805711 104,5385008 102,5497195 112,3151453
91,40744518 0 80,64762876 122,0805711 104,5385008 102,5497195
99,51555329 0 91,40744518 80,64762876 122,0805711 104,5385008
106,527282 1 99,51555329 91,40744518 80,64762876 122,0805711
98,49566548 0 106,527282 99,51555329 91,40744518 80,64762876
106,7567568 1 98,49566548 106,527282 99,51555329 91,40744518
|