Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_chi_squared_tests.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Chi-Squared Test, McNemar Test, and Fisher Exact Test | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Fri, 21 Dec 2012 09:21:56 -0500 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2012/Dec/21/t1356100166t557x0xtn4vwn8q.htm/, Retrieved Thu, 31 Oct 2024 23:09:56 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=203718, Retrieved Thu, 31 Oct 2024 23:09:56 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 120 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Chi-Squared and McNemar Tests] [] [2010-11-16 14:33:59] [b98453cac15ba1066b407e146608df68] - R PD [Chi-Squared Test, McNemar Test, and Fisher Exact Test] [Paper 2012, deel ...] [2012-12-21 14:21:56] [e4c351aee2a0bb2c047702ea90f356fa] [Current] - R P [Chi-Squared Test, McNemar Test, and Fisher Exact Test] [Paper 2012, deel ...] [2012-12-21 15:11:39] [346d881f0ddec1e86442b342eb3e8104] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
4 'Yes' 'Treatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'Treatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'Yes' 'Treatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'Yes' 'Bad' 4 'Yes' 'Treatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'Yes' 'Good' 4 'No' 'Treatment' NA 'UsedStats' 'No' 'Yes' 'Good' 4 'Yes' 'Treatment' NA 'UsedStats' 'Yes' 'Yes' 'Bad' 4 'Yes' 'Treatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'Treatment' NA 'UsedStats' 'Yes' 'Yes' 'Good' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Bad' 4 'Yes' 'NoTreatment' NA 'UsedStats' 'No' 'Yes' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'No' 'Treatment' NA 'UsedStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'Yes' 'Bad' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Bad' 4 'No' 'Treatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'Yes' 'Treatment' NA 'UsedStats' 'No' 'Yes' 'Bad' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'No' 'Treatment' NA 'NoStats' 'No' 'Yes' 'Bad' 4 'No' 'NoTreatment' NA 'UsedStats' 'Yes' 'Yes' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'No' 'Good' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'Yes' 'Treatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'Treatment' NA 'UsedStats' 'No' 'No' 'Bad' 4 'Yes' 'Treatment' NA 'UsedStats' 'Yes' 'Yes' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'Yes' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'Treatment' NA 'UsedStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'Yes' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'Yes' 'Treatment' NA 'UsedStats' 'Yes' 'Yes' 'Good' 4 'Yes' 'Treatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'Yes' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'Yes' 'Treatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'Treatment' NA 'UsedStats' 'Yes' 'Yes' 'Bad' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'No' 'Good' 4 'Yes' 'NoTreatment' NA 'UsedStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'Treatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'UsedStats' 'No' 'Yes' 'Good' 4 'No' 'Treatment' NA 'UsedStats' 'Yes' 'No' 'Good' 4 'No' 'Treatment' NA 'NoStats' 'No' 'Yes' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'Yes' 'NoTreatment' NA 'UsedStats' 'No' 'No' 'Good' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'UsedStats' 'Yes' 'No' 'Bad' 4 'No' 'NoTreatment' NA 'NoStats' 'No' 'Yes' 'Good' 4 'Yes' 'NoTreatment' NA 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'Yes' NA 'Treatment' 'UsedStats' 'No' 'No' 'Good' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'Yes' 'Bad' 2 'Yes' NA 'Treatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'Yes' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'Treatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'Yes' NA 'Treatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'Treatment' 'UsedStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'Treatment' 'UsedStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'Treatment' 'UsedStats' 'No' 'Yes' 'Bad' 2 'No' NA 'Treatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'UsedStats' 'No' 'No' 'Bad' 2 'Yes' NA 'Treatment' 'UsedStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'Treatment' 'UsedStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'UsedStats' 'No' 'Yes' 'Good' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'No' NA 'Treatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'Yes' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'Yes' NA 'NoTreatment' 'UsedStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'UsedStats' 'No' 'Yes' 'Good' 2 'Yes' NA 'Treatment' 'UsedStats' 'No' 'Yes' 'Good' 2 'No' NA 'Treatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'UsedStats' 'Yes' 'No' 'Good' 2 'No' NA 'Treatment' 'UsedStats' 'No' 'No' 'Good' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'Yes' 'Good' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'Yes' 'Bad' 2 'No' NA 'Treatment' 'NoStats' 'No' 'No' 'Good' 2 'No' NA 'Treatment' 'UsedStats' 'No' 'No' 'Bad' 2 'No' NA 'Treatment' 'NoStats' 'No' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Bad' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'Yes' 'Good' 2 'No' NA 'NoTreatment' 'NoStats' 'No' 'No' 'Good' 2 'Yes' NA 'NoTreatment' 'UsedStats' 'Yes' 'No' 'Bad' 2 'Yes' NA 'NoTreatment' 'UsedStats' 'Yes' 'Yes' 'Bad' 2 'Yes' NA 'NoTreatment' 'UsedStats' 'No' 'No' 'Bad' | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
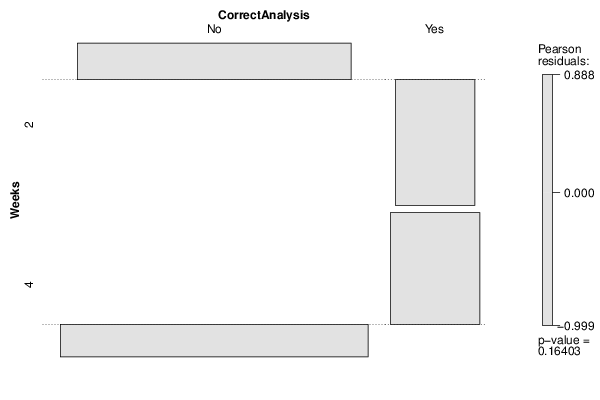
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = 6 ; par3 = Pearson Chi-Squared ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = 6 ; par3 = Pearson Chi-Squared ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
library(vcd) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||