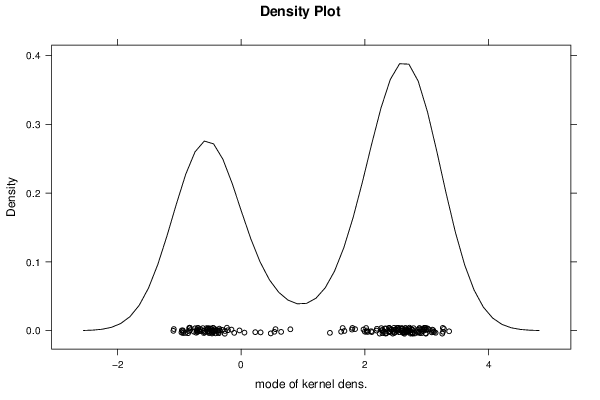
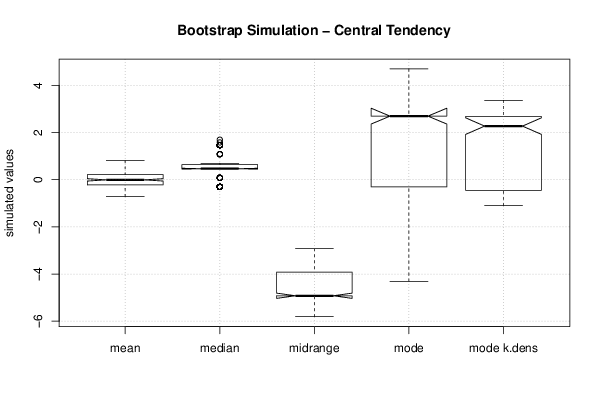
1,46429
2,46429
2,46429
-1,53571
3,46429
-7,53571
0,464286
2,46429
1,46429
-0,535714
2,46429
-4,53571
0,464286
-0,535714
-1,53571
-4,53571
0,464286
1,46429
1,46429
-4,53571
-3,53571
3,46429
1,46429
-1,53571
5,46429
-10,5357
10,4643
0,464286
3,46429
-3,53571
-3,53571
-0,535714
5,46429
-1,53571
3,46429
1,46429
-9,53571
-5,53571
2,46429
6,46429
3,46429
3,46429
4,46429
4,46429
-1,53571
3,46429
-4,53571
-0,535714
-3,53571
5,46429
3,46429
-2,53571
-0,535714
1,46429
-1,53571
7,46429
1,46429
-6,53571
-11,5357
9,46429
8,46429
3,46429
1,46429
-0,535714
-0,535714
0,464286
-1,53571
-0,535714
-2,53571
-0,535714
5,46429
-0,535714
2,46429
3,46429
-5,53571
-3,53571
4,46429
0,464286
-7,53571
4,46429
2,46429
-7,53571
2,46429
0,464286
-9,53571
3,46429
-2,53571
2,46429
4,46429
-1,53571
1,46429
0,464286
0,464286
2,46429
-0,535714
0,464286
6,46429
3,46429
4,46429
1,46429
1,46429
-0,535714
-11,5357
-2,53571
0,464286
-8,53571
-11,5357
-4,53571
-1,53571
-1,53571
-0,535714
-0,535714
2,69277
1,69277
0,692771
4,69277
9,69277
-3,30723
6,69277
2,69277
2,69277
-2,30723
-2,30723
2,69277
-1,30723
-5,30723
-0,307229
-4,30723
3,69277
4,69277
4,69277
-1,30723
-1,30723
-4,30723
-1,30723
-1,30723
2,69277
0,692771
1,69277
-1,30723
-0,307229
-0,307229
-17,3072
2,69277
2,69277
-0,307229
-5,30723
-4,30723
-13,3072
3,69277
-3,30723
3,69277
3,69277
-1,30723
4,69277
-0,307229
7,69277
2,69277
6,69277
-2,30723
7,69277
0,692771
-1,30723
2,69277
6,69277
1,69277
7,69277
4,69277
0,692771
1,69277
7,69277
-0,307229
8,69277
4,69277
4,69277
-0,307229
-0,307229
-4,30723
-0,307229
-0,307229
2,69277
-2,30723
4,69277
-2,30723
-1,30723
4,69277
4,69277
4,69277
3,69277
-1,30723
5,69277
-10,3072
-3,30723
-7,30723
-3,30723
9,69277
4,69277
-16,3072
-4,30723
0,692771
2,69277
1,69277
-3,30723
-0,307229
-0,307229
1,69277
-4,30723
2,69277
-20,3072
-2,30723
4,69277
2,69277
-8,30723
-2,30723
3,69277
-9,30723
-2,30723
2,69277
3,69277
8,69277
-2,30723
-5,30723
8,69277
-4,30723
-1,30723
1,69277
-4,30723
2,69277
2,69277
-1,30723
-16,3072
-0,307229
3,69277
-0,307229
-16,3072
3,69277
1,69277
-4,30723
-17,3072
-5,30723
3,69277
-3,30723
-0,307229
6,69277
5,69277
2,69277
-3,30723
-0,307229
1,69277
-1,30723
3,69277
-1,30723
2,69277
-5,30723
6,69277
5,69277
1,69277
1,69277
-2,30723
-5,30723
1,69277
6,69277
-10,3072
-0,307229
-3,30723
2,69277
-1,30723
-7,30723
6,69277
2,69277
-4,30723
4,69277
-18,3072
5,69277
-0,307229
2,69277
1,69277
3,69277 |