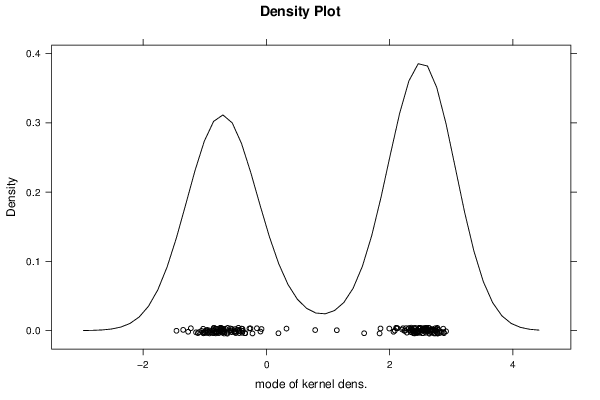
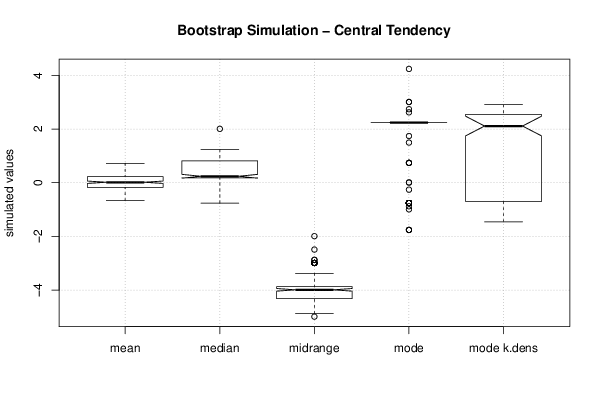
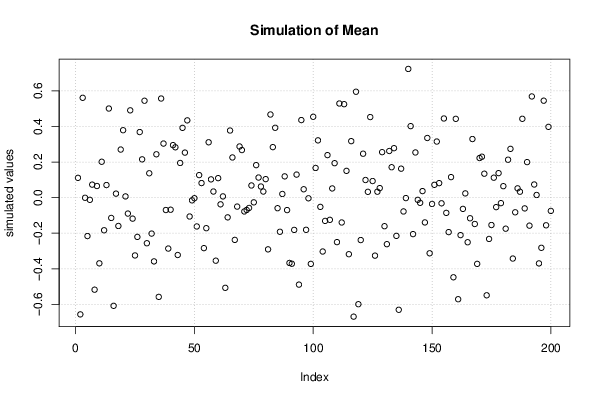
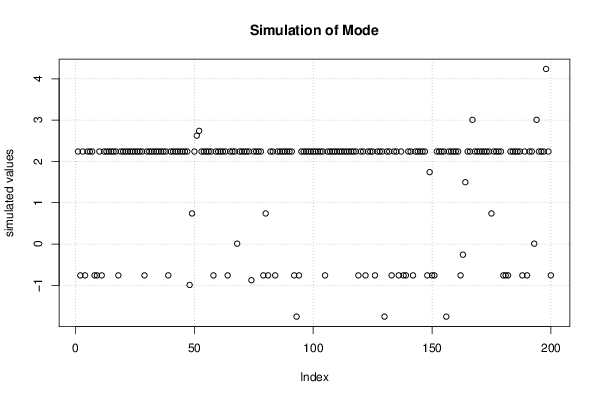
2,00833
1,24051
3,00833
-2,75949
2,24051
-8,75949
1,00833
1,24051
0,240506
-1,75949
1,24051
-5,75949
-0,759494
0,00833333
-0,991667
-3,99167
-0,759494
2,00833
0,240506
-3,99167
-4,75949
2,24051
2,00833
-2,75949
4,24051
-11,7595
9,24051
1,00833
2,24051
-2,99167
-2,99167
0,00833333
4,24051
-2,75949
2,24051
0,240506
-8,99167
-6,75949
1,24051
7,00833
2,24051
2,24051
3,24051
3,24051
-2,75949
4,00833
-5,75949
-1,75949
-2,99167
4,24051
2,24051
-3,75949
-1,75949
0,240506
-2,75949
6,24051
2,00833
-7,75949
-10,9917
8,24051
7,24051
4,00833
2,00833
-1,75949
0,00833333
-0,759494
-0,991667
-1,75949
-3,75949
0,00833333
6,00833
0,00833333
3,00833
2,24051
-4,99167
-2,99167
3,24051
1,00833
-6,99167
3,24051
3,00833
-6,99167
3,00833
-0,759494
-8,99167
2,24051
-3,75949
3,00833
5,00833
-0,991667
0,240506
-0,759494
-0,759494
3,00833
-1,75949
1,00833
5,24051
2,24051
3,24051
0,240506
0,240506
0,00833333
-12,7595
-3,75949
-0,759494
-7,99167
-10,9917
-3,99167
-0,991667
-0,991667
0,00833333
-1,75949
2,24051
1,24051
0,240506
4,24051
11,0083
-3,75949
6,24051
4,00833
2,24051
-0,991667
-0,991667
2,24051
-1,75949
-5,75949
-0,759494
-4,75949
3,24051
4,24051
4,24051
0,00833333
-1,75949
-4,75949
-1,75949
-1,75949
2,24051
0,240506
3,00833
-1,75949
-0,759494
-0,759494
-17,7595
2,24051
4,00833
1,00833
-5,75949
-2,99167
-11,9917
3,24051
-1,99167
3,24051
3,24051
0,00833333
4,24051
-0,759494
7,24051
4,00833
8,00833
-0,991667
9,00833
0,240506
0,00833333
2,24051
8,00833
1,24051
9,00833
4,24051
2,00833
3,00833
7,24051
1,00833
8,24051
4,24051
4,24051
-0,759494
-0,759494
-2,99167
-0,759494
1,00833
4,00833
-0,991667
4,24051
-0,991667
-1,75949
6,00833
6,00833
6,00833
5,00833
-1,75949
5,24051
-10,7595
-3,75949
-5,99167
-1,99167
9,24051
6,00833
-14,9917
-2,99167
2,00833
2,24051
1,24051
-1,99167
1,00833
-0,759494
3,00833
-4,75949
2,24051
-18,9917
-2,75949
4,24051
2,24051
-6,99167
-0,991667
5,00833
-9,75949
-2,75949
2,24051
3,24051
10,0083
-0,991667
-3,99167
8,24051
-4,75949
0,00833333
3,00833
-2,99167
2,24051
2,24051
0,00833333
-14,9917
1,00833
3,24051
-0,759494
-16,7595
3,24051
3,00833
-4,75949
-17,7595
-5,75949
5,00833
-1,99167
-0,759494
8,00833
5,24051
2,24051
-1,99167
-0,759494
3,00833
-1,75949
3,24051
0,00833333
2,24051
-3,99167
6,24051
7,00833
1,24051
3,00833
-0,991667
-5,75949
1,24051
8,00833
-10,7595
-0,759494
-1,99167
2,24051
0,00833333
-5,99167
6,24051
2,24051
-2,99167
4,24051
-16,9917
7,00833
-0,759494
4,00833
3,00833
3,24051 |