Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_bootstrapplot1.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
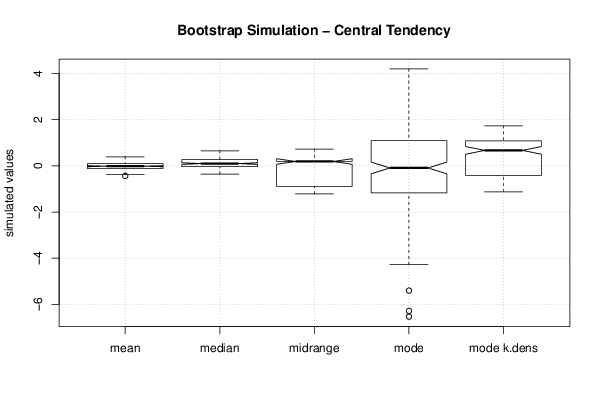
| Title produced by software | Bootstrap Plot - Central Tendency | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Sat, 13 Dec 2014 12:30:20 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2014/Dec/13/t1418473911qqam1kjq66o6pf0.htm/, Retrieved Thu, 16 May 2024 21:20:37 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=267032, Retrieved Thu, 16 May 2024 21:20:37 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 106 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Bootstrap Plot - Central Tendency] [] [2014-11-04 10:49:59] [32b17a345b130fdf5cc88718ed94a974] - R D [Bootstrap Plot - Central Tendency] [] [2014-11-12 15:33:27] [d253a55552bf9917a397def3be261e30] - D [Bootstrap Plot - Central Tendency] [] [2014-12-13 12:30:20] [940a3d9bc049bdd1effc6e8b1116301d] [Current] - D [Bootstrap Plot - Central Tendency] [] [2014-12-14 13:00:35] [d253a55552bf9917a397def3be261e30] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
0.932222 -3.40718 2.02093 1.36824 -3.79056 -3.38055 -1.23504 3.31485 3.26324 -0.162007 -4.14466 -0.0353275 -5.40106 1.74394 -0.982709 -0.310041 1.91354 -0.325889 -0.891366 -2.14726 0.791716 -3.57529 2.88177 -2.73686 1.69897 0.900939 -3.62329 2.53661 3.31244 -0.603295 0.719479 -0.824002 0.425903 -4.314 -0.827149 -2.52479 1.55106 0.928113 -0.028673 1.74671 -2.28063 -0.527203 -3.45186 -0.169304 1.7191 -0.439106 3.83894 -1.61396 2.23742 -2.01029 4.19365 2.82941 0.87747 -1.05408 0.0306141 0.297232 2.82935 0.130906 -5.45826 4.74926 0.387721 2.65825 -2.69075 1.48456 -0.599803 2.58118 -2.44582 0.583253 -1.7366 -0.931429 2.46733 1.39476 -0.71745 1.43063 -0.856571 3.77843 -4.06047 1.74959 0.903734 1.95243 0.783888 0.324938 -1.66152 0.120254 3.16781 0.392802 -4.71512 -0.763504 1.02922 1.99095 2.11744 -1.09754 2.04121 3.91196 -4.33172 -1.18583 -1.20406 -2.32271 -1.32617 -3.74664 2.09148 3.67782 -2.24472 1.88483 -0.00112466 -0.559093 -1.10596 4.06807 -1.51748 3.86307 -4.82211 -1.81613 -0.670688 0.986877 1.79823 -6.13285 0.572984 3.02791 2.36338 -1.29867 1.30845 -3.13411 1.86285 -2.27486 2.066 1.25583 4.73285 -3.2583 -0.604271 1.2631 -2.04116 1.74526 1.25777 1.72918 -1.84841 1.11176 0.923519 0.269473 -0.636317 3.25515 -1.69528 1.4904 0.0861235 -0.204194 3.294 0.79281 2.87255 0.860894 0.265504 1.31237 -0.224824 -2.44669 0.15643 -1.19531 1.80645 -5.89349 1.35698 2.23385 -0.779467 1.36121 -0.612783 1.7998 1.09944 -2.84872 -2.49653 2.2386 0.508086 0.281476 6.90427 1.28433 -0.267105 1.46841 1.88506 -1.00054 -3.13529 0.778232 2.60497 -1.46431 -0.616958 -0.567452 2.72638 -0.932362 2.23324 -0.847608 -3.67248 -2.27175 -2.0986 2.3869 -0.0399423 -1.94751 0.437445 2.793 -1.84805 -1.67959 -0.145587 -2.57096 1.47757 -2.89814 3.2941 -0.537296 0.416 -2.11153 -0.190319 1.21825 0.950062 0.673744 0.372071 -1.07387 -0.478571 -0.12629 1.35631 -1.43241 0.440395 -0.797422 2.43008 1.65594 -0.383124 0.78967 -2.58556 -1.38788 -0.684021 -0.616442 1.34645 -1.24077 2.22666 -1.37847 -1.20355 1.99146 -4.27317 -0.773728 3.03373 2.60617 2.86588 2.76515 1.65633 3.63065 3.32698 3.51234 2.29815 -0.0894453 -2.15762 -2.02724 -6.27872 0.135422 -1.42627 -1.04298 -0.625054 -1.15017 -2.18658 0.754245 1.95562 4.1108 -2.5743 -1.1334 0.565461 0.942532 0.934201 -0.33767 -1.97428 -1.62172 -0.13279 1.19369 -1.64225 0.201401 -4.23566 -5.46212 -1.82565 -6.5295 -0.932446 -1.84111 -3.74594 1.18795 1.21946 -0.0542749 -0.669771 1.62901 2.03339 -1.02664 0.854178 -0.661109 0.620873 0.938601 0.513566 0.00438837 0.732725 -2.63675 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
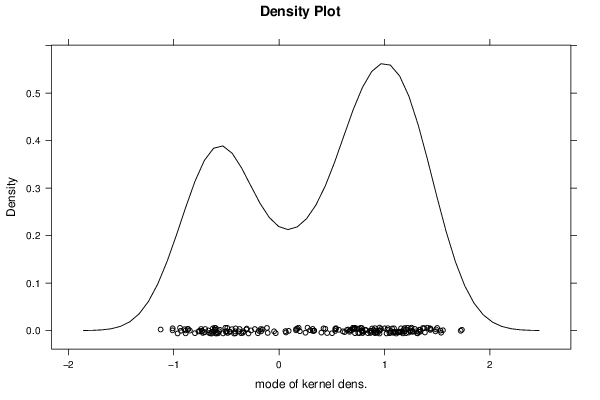
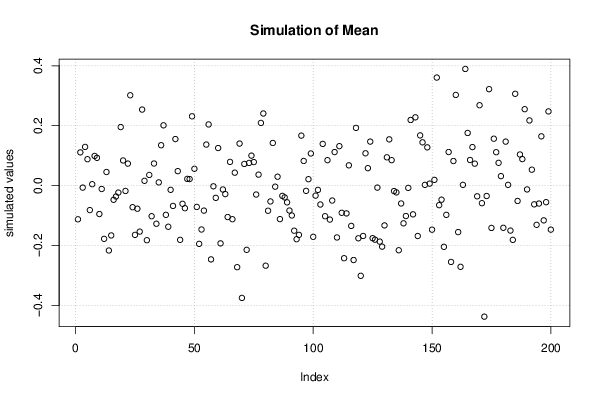
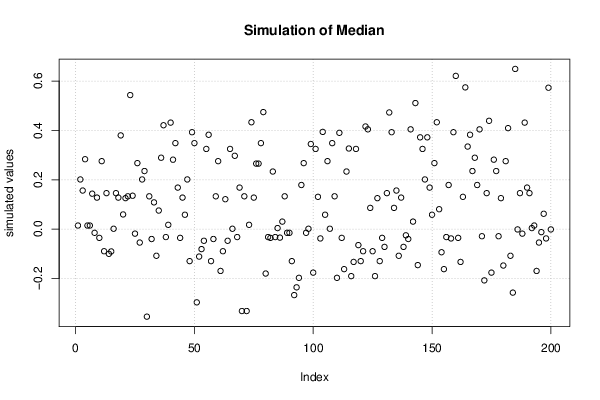
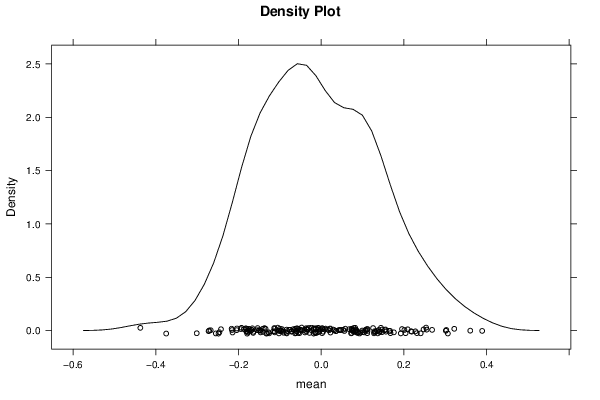
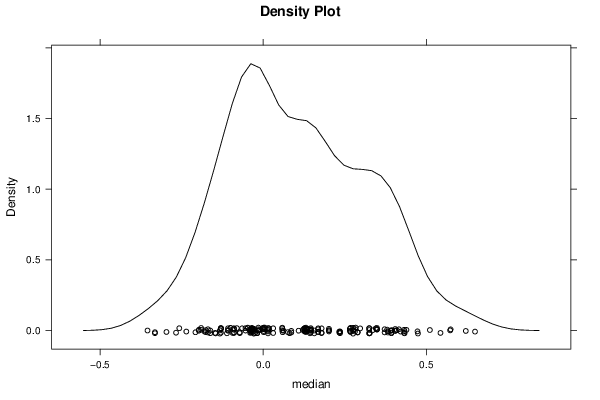
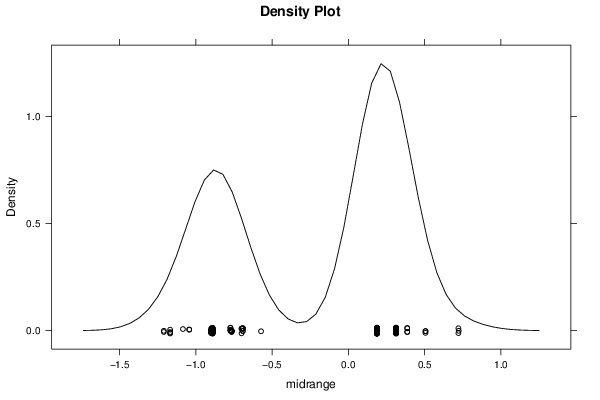
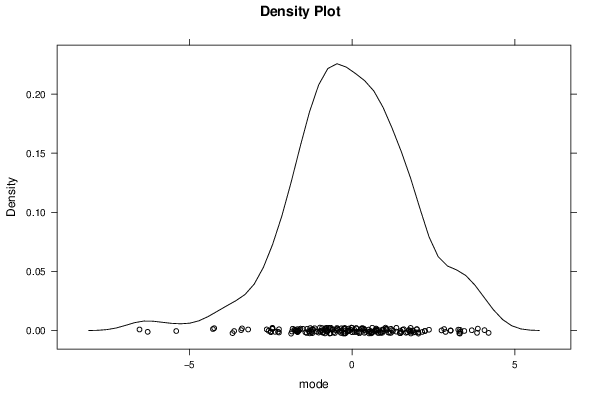
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 200 ; par2 = 5 ; par3 = 0 ; par4 = P1 P5 Q1 Q3 P95 P99 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 200 ; par2 = 5 ; par3 = 0 ; par4 = P1 P5 Q1 Q3 P95 P99 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||