Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_Simple Regression Y ~ X.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Simple Linear Regression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Sun, 14 Dec 2014 12:55:18 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2014/Dec/14/t1418561733yj2so3na6mci9u5.htm/, Retrieved Thu, 16 May 2024 22:09:33 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=267528, Retrieved Thu, 16 May 2024 22:09:33 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 128 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Blocked Bootstrap Plot - Central Tendency] [] [2014-11-02 13:37:17] [cc401d1001c65f55a3dfc6f2420e9570] - RMPD [Simple Linear Regression] [] [2014-11-02 15:26:26] [cc401d1001c65f55a3dfc6f2420e9570] - RM [Simple Linear Regression] [] [2014-11-05 18:55:35] [e296091fd6311efcd9175c015e8e9c4e] - MPD [Simple Linear Regression] [] [2014-12-09 12:47:37] [36c866d94170840abc594fd3e7d5794f] - PD [Simple Linear Regression] [] [2014-12-14 12:50:11] [36c866d94170840abc594fd3e7d5794f] - D [Simple Linear Regression] [] [2014-12-14 12:55:18] [72ee53c6f28232e74174360ca89644de] [Current] - D [Simple Linear Regression] [] [2014-12-14 12:59:02] [36c866d94170840abc594fd3e7d5794f] - D [Simple Linear Regression] [] [2014-12-14 13:02:17] [36c866d94170840abc594fd3e7d5794f] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
18 12.9 31 12.2 39 12.8 46 7.4 31 6.7 67 12.6 35 14.8 52 13.3 77 11.1 37 8.2 32 11.4 36 6.4 38 10.6 69 12 21 6.3 26 11.3 54 11.9 36 9.3 42 9.6 23 10 34 6.4 112 13.8 35 10.8 47 13.8 47 11.7 37 10.9 109 16.1 24 13.4 20 9.9 22 11.5 23 8.3 32 11.7 30 9 92 9.7 43 10.8 55 10.3 16 10.4 49 12.7 71 9.3 43 11.8 29 5.9 56 11.4 46 13 19 10.8 23 12.3 59 11.3 30 11.8 61 7.9 7 12.7 38 12.3 32 11.6 16 6.7 19 10.9 22 12.1 48 13.3 23 10.1 26 5.7 33 14.3 9 8 24 13.3 34 9.3 48 12.5 18 7.6 43 15.9 33 9.2 28 9.1 71 11.1 26 13 67 14.5 34 12.2 80 12.3 29 11.4 16 8.8 59 14.6 32 12.6 43 13 38 12.6 29 13.2 36 9.9 32 7.7 35 10.5 21 13.4 29 10.9 12 4.3 37 10.3 37 11.8 47 11.2 51 11.4 32 8.6 21 13.2 13 12.6 14 5.6 -2 9.9 20 8.8 24 7.7 11 9 23 7.3 24 11.4 14 13.6 52 7.9 15 10.7 23 10.3 19 8.3 35 9.6 24 14.2 39 8.5 29 13.5 13 4.9 8 6.4 18 9.6 24 11.6 19 11.1 23 4.35 16 12.7 33 18.1 32 17.85 37 16.6 14 12.6 52 17.1 75 19.1 72 16.1 15 13.35 29 18.4 13 14.7 40 10.6 19 12.6 24 16.2 121 13.6 93 18.9 36 14.1 23 14.5 85 16.15 41 14.75 46 14.8 18 12.45 35 12.65 17 17.35 4 8.6 28 18.4 44 16.1 10 11.6 38 17.75 57 15.25 23 17.65 36 16.35 22 17.65 40 13.6 31 14.35 11 14.75 38 18.25 24 9.9 37 16 37 18.25 22 16.85 15 14.6 2 13.85 43 18.95 31 15.6 29 14.85 45 11.75 25 18.45 4 15.9 31 17.1 -4 16.1 66 19.9 61 10.95 32 18.45 31 15.1 39 15 19 11.35 31 15.95 36 18.1 42 14.6 21 15.4 21 15.4 25 17.6 32 13.35 26 19.1 28 15.35 32 7.6 41 13.4 29 13.9 33 19.1 17 15.25 13 12.9 32 16.1 30 17.35 34 13.15 59 12.15 13 12.6 23 10.35 10 15.4 5 9.6 31 18.2 19 13.6 32 14.85 30 14.75 25 14.1 48 14.9 35 16.25 67 19.25 15 13.6 22 13.6 18 15.65 33 12.75 46 14.6 24 9.85 14 12.65 12 19.2 38 16.6 12 11.2 28 15.25 41 11.9 12 13.2 31 16.35 33 12.4 34 15.85 21 18.15 20 11.15 44 15.65 52 17.75 7 7.65 29 12.35 11 15.6 26 19.3 24 15.2 7 17.1 60 15.6 13 18.4 20 19.05 52 18.55 28 19.1 25 13.1 39 12.85 9 9.5 19 4.5 13 11.85 60 13.6 19 11.7 34 12.4 14 13.35 17 11.4 45 14.9 66 19.9 48 11.2 29 14.6 -2 17.6 51 14.05 2 16.1 24 13.35 40 11.85 20 11.95 19 14.75 16 15.15 20 13.2 40 16.85 27 7.85 25 7.7 49 12.6 39 7.85 61 10.95 19 12.35 67 9.95 45 14.9 30 16.65 8 13.4 19 13.95 52 15.7 22 16.85 17 10.95 33 15.35 34 12.2 22 15.1 30 17.75 25 15.2 38 14.6 26 16.65 13 8.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
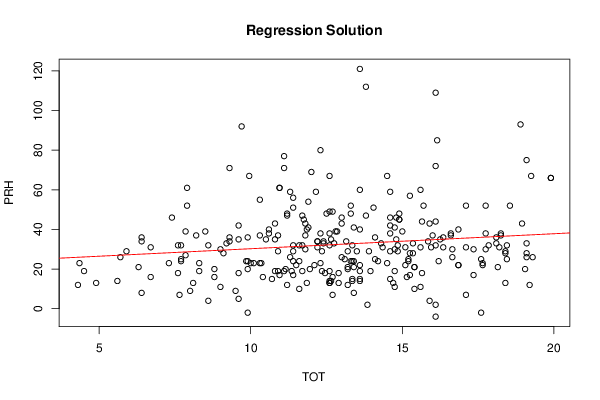
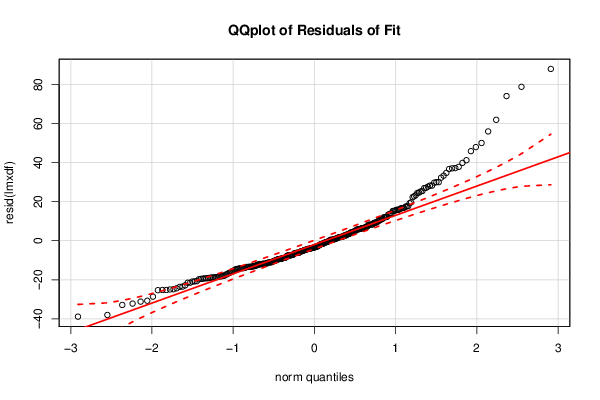
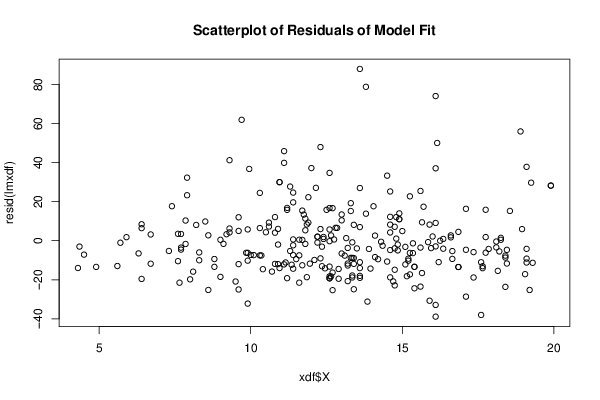
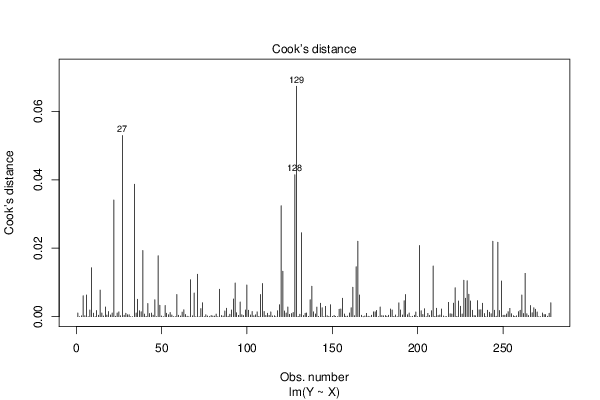
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = 2 ; par3 = TRUE ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = 2 ; par3 = TRUE ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
cat1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||