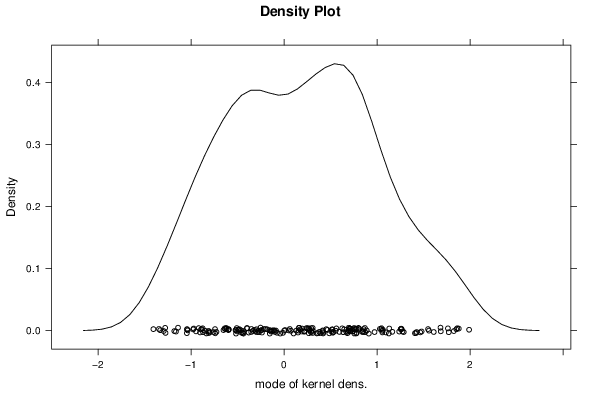
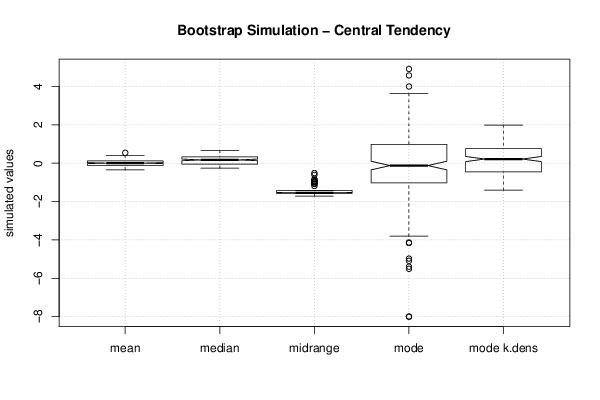
1,344
0,994
1,205
-4,332
-4,163
-1,390
2,907
3,371
-0,737
-4,134
-0,874
-5,399
0,203
-0,427
-1,527
2,123
-0,162
-1,384
-1,524
-0,253
-5,090
2,271
-3,485
1,021
-0,047
-3,703
4,236
3,999
-1,721
-0,045
-1,063
-0,185
-2,225
-2,159
0,715
0,594
-0,764
2,049
-2,843
-0,438
-4,269
-0,213
0,899
-1,524
4,239
-1,772
1,942
-1,273
3,114
2,999
1,108
-0,921
-0,852
0,103
2,673
-0,837
-5,158
3,495
0,662
3,255
-3,148
0,415
-2,091
2,430
-3,543
0,673
-1,575
-0,877
1,717
2,449
-0,448
0,786
-0,185
4,914
1,653
1,131
2,435
1,105
1,531
-2,531
0,943
3,549
1,540
-5,231
0,177
1,785
2,490
2,638
-0,829
2,406
3,806
-4,717
-1,328
-1,061
-2,055
-1,462
-3,802
1,550
3,625
-2,079
1,979
0,476
-0,257
-0,968
4,575
-0,706
4,438
-4,372
-1,562
0,150
1,319
1,512
-7,752
0,303
1,988
1,355
1,755
-0,022
-1,178
2,854
-0,213
0,497
2,341
2,559
-3,557
-1,966
0,426
-0,239
3,616
-0,224
-0,338
-0,254
0,217
0,540
-0,791
-0,851
2,045
-3,544
2,138
1,268
-1,492
1,404
0,167
2,075
0,253
1,855
-0,896
0,129
-0,620
1,043
-6,851
1,294
1,728
-0,336
1,193
-0,058
2,010
0,749
-0,328
-1,298
3,459
1,203
0,435
4,863
3,346
-1,020
3,078
1,164
0,695
-2,264
1,333
3,546
-1,258
-1,897
-1,897
3,163
-1,371
2,698
-0,335
-4,652
-0,341
-0,890
2,537
1,041
-2,858
1,434
2,747
-1,530
-2,224
-0,140
-2,500
1,489
-3,222
4,055
0,491
1,133
-1,700
1,085
2,051
1,853
0,553
0,599
-1,909
0,563
-2,105
2,012
-2,102
0,171
4,724
2,601
-0,735
-0,920
-2,078
-0,877
-0,098
-1,392
2,081
0,862
-1,441
0,617
2,016
-4,651
-2,525
0,370
2,550
2,619
1,119
2,054
3,332
2,937
2,835
3,443
-0,671
-3,695
-3,734
-8,002
-0,458
-1,974
-0,422
-0,718
-1,520
-1,418
0,961
3,346
-2,484
-0,318
0,990
-0,578
0,621
-2,036
-3,906
-1,593
0,547
2,023
-3,767
2,058
-4,720
-6,613
-1,253
-5,940
-1,020
-1,588
-3,267
0,961
2,603
0,275
0,480
1,882
1,866
-1,488
0,840
-0,953
2,144
2,888
1,006
-0,257
1,633
-3,750
|