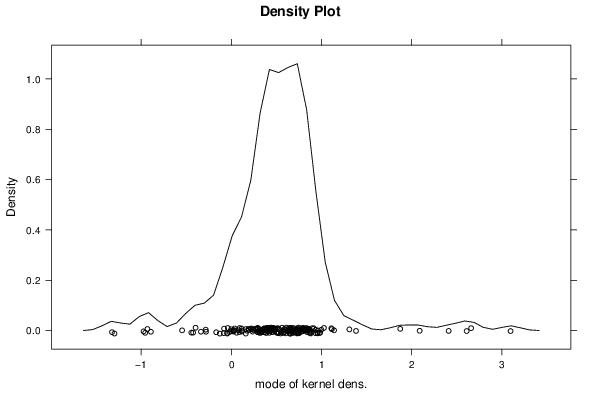
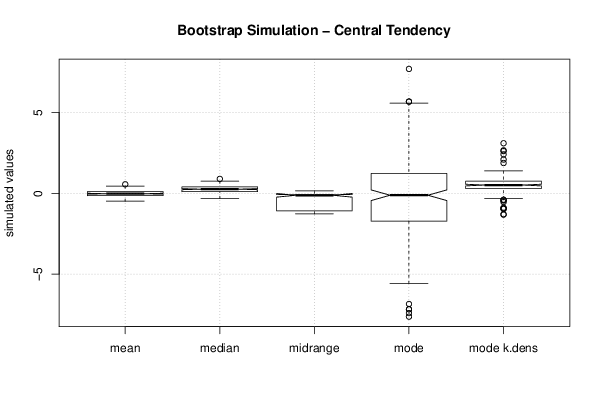
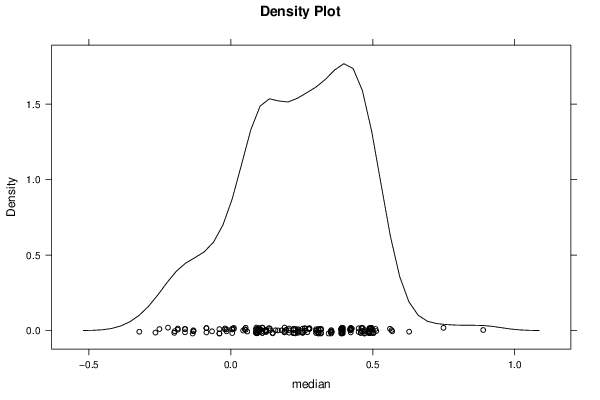
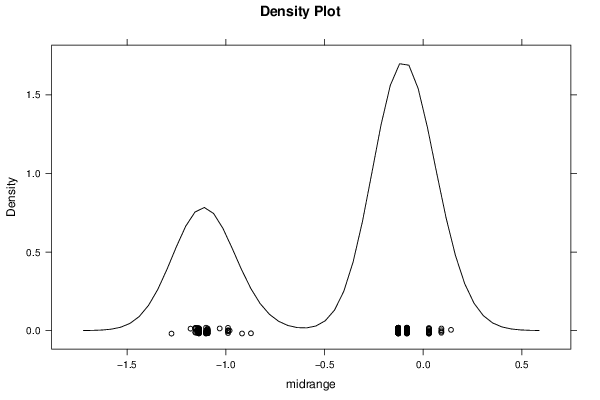
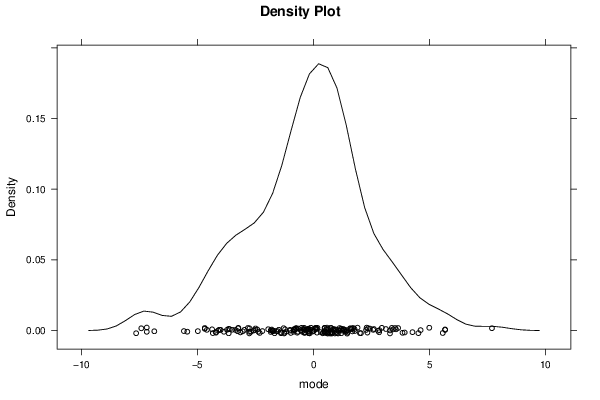
-1,29552
-6,41697
-1,61397
-1,432
-6,30415
-7,18061
-2,50838
1,33138
-0,185791
-2,39986
-7,42168
-2,67362
-7,5121
-1,66185
-2,01918
-4,68132
-0,477008
-2,01089
-3,64904
-4,19613
-2,1094
-7,00176
-0,260993
-4,36442
-0,402389
-2,31204
-4,11383
1,6823
0,842372
-3,99359
-1,39474
-3,23674
-2,41574
-6,99811
-4,25033
-4,01933
-1,71821
-2,51649
-2,45004
-0,413312
-5,5793
-1,74536
-7,63817
-2,749
-1,06748
-2,51022
1,15666
-3,65444
-0,979399
-4,37096
0,774192
-0,210468
-0,970903
-4,3685
-2,8245
-1,498
-0,0403176
-2,9682
-6,79938
1,28564
-2,62465
0,300274
-5,38461
-1,48788
-4,19714
0,891956
-5,60067
-2,71173
-3,14672
-2,43686
0,0875358
-0,24235
-2,83652
-1,73362
-3,59933
1,07464
-6,85434
0,450215
-0,548117
-0,537537
-0,853124
-1,18345
-4,97802
-1,98526
1,38977
-1,00532
-7,86149
-2,51183
-1,2259
-0,833874
-0,137417
-3,85482
0,470952
0,488184
-6,95483
-3,5232
-3,42125
-4,95551
-3,8284
-6,24852
-1,14494
1,08723
-4,66607
-1,00529
-1,77136
-2,65774
-3,21353
1,8842
-3,72081
1,24278
-6,66835
-4,8953
-2,97454
-1,21016
-1,39683
-7,94904
1,24221
4,52786
3,93632
2,78777
0,457843
1,3429
4,65396
1,86028
1,7369
3,94764
3,88628
-1,98633
0,252967
2,95037
0,886392
5,23545
1,1226
1,13831
2,00667
2,32906
2,29742
0,622541
0,110985
4,10634
-2,00977
4,83695
2,22021
-0,451989
4,02805
2,53262
4,99623
2,13408
2,6064
3,2582
-0,22163
-1,62235
1,52565
1,94623
3,32681
-3,35521
2,52814
3,80448
2,69972
2,19382
1,11372
3,81633
2,88411
0,926709
-0,000846216
5,02158
3,84178
2,63474
5,30497
4,41108
-0,557277
5,39836
2,44693
1,93001
-0,805945
3,12845
5,0224
0,724264
0,632938
0,749219
5,3486
-0,132022
5,66036
1,69896
-3,45418
0,495473
0,761351
5,17267
3,09294
-0,548312
2,7708
4,2205
0,0936131
-1,17853
1,46968
-1,67149
3,65109
-2,44933
5,67622
1,41517
1,33821
1,20204
2,61317
3,03466
3,13758
1,00981
0,778034
0,390412
3,15852
0,50974
3,09872
-0,844599
0,317464
0,486873
7,69656
4,66444
-0,18824
1,61265
0,0102794
2,07927
2,8454
-0,374375
3,41705
0,105157
3,91895
-0,270727
2,27793
4,90667
-2,76196
-1,29698
3,09531
5,57596
3,53736
3,7908
3,30151
5,64045
5,59128
5,13472
5,4585
1,28149
-0,81367
-1,83786
-7,41442
0,866838
0,48938
-0,676063
0,514872
0,389144
1,06188
2,12067
5,04962
4,08731
-1,7065
2,24683
3,52175
0,850088
2,95909
0,157149
-1,52637
0,137609
2,35835
3,51591
-1,07826
3,65263
-3,49834
-4,049
0,39459
-5,00293
-1,57423
0,22129
-2,63825
2,04925
4,28113
1,18677
1,47136
3,78654
3,45938
-0,5734
2,97713
0,634013
4,05617
4,82513
2,44126
0,992056
3,84166
-2,91006
|