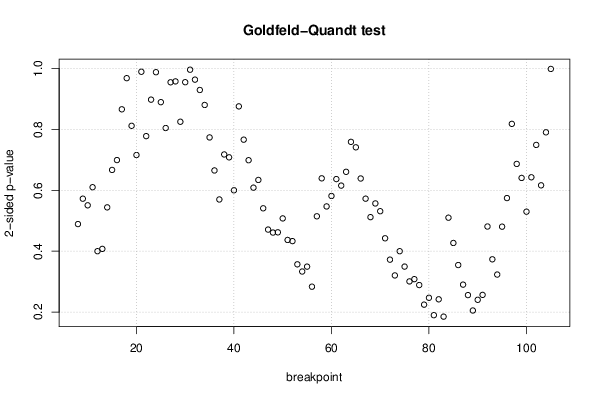
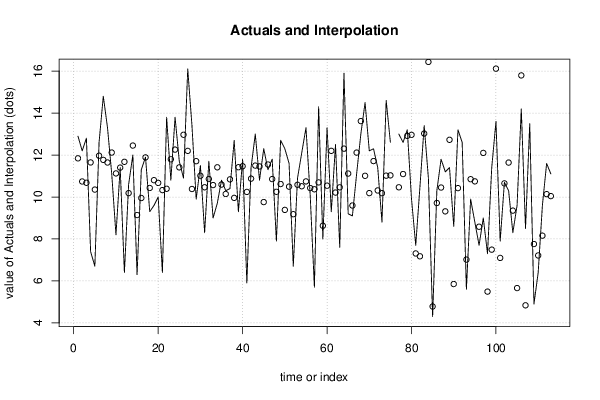
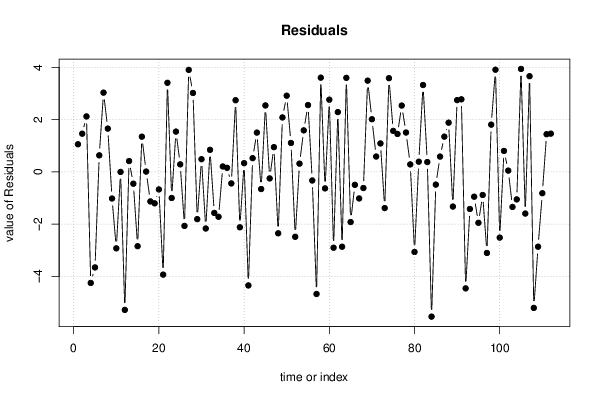
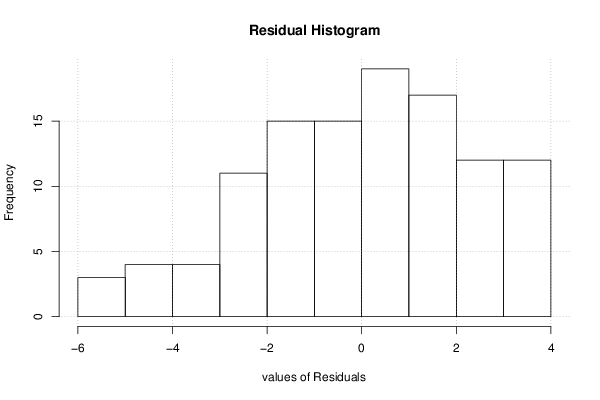
0,67 0,67 149 68 12,9
0,83 0,33 139 39 12,2
1,00 0,67 148 32 12,8
0,83 0,00 158 62 7,4
0,67 0,00 128 33 6,7
0,00 0,00 224 52 12,6
0,83 0,67 159 62 14,8
0,50 0,67 105 77 13,3
0,83 0,00 159 76 11,1
0,33 0,67 167 41 8,2
0,50 1,00 165 48 11,4
0,67 0,00 159 63 6,4
1,00 0,00 119 30 10,6
0,50 0,67 176 78 12,0
0,67 0,33 54 19 6,3
0,17 0,67 91 31 11,3
0,83 0,33 163 66 11,9
0,67 0,33 124 35 9,3
0,67 0,33 137 42 9,6
0,67 0,00 121 45 10,0
0,50 0,67 153 21 6,4
1,00 0,33 148 25 13,8
0,83 0,33 221 44 10,8
0,83 0,33 188 69 13,8
1,00 0,67 149 54 11,7
0,67 0,00 244 74 10,9
1,00 0,33 148 80 16,1
0,83 0,67 92 42 13,4
1,00 1,00 150 61 9,9
0,83 0,67 153 41 11,5
0,67 0,33 94 46 8,3
0,67 0,00 156 39 11,7
1,00 0,67 132 34 9,0
0,67 0,67 161 51 9,7
1,00 1,00 105 42 10,8
1,00 1,00 97 31 10,3
0,50 0,33 151 39 10,4
0,67 0,00 131 20 12,7
0,83 0,67 166 49 9,3
1,00 0,67 157 53 11,8
1,00 0,67 111 31 5,9
1,00 0,67 145 39 11,4
1,00 0,33 162 54 13,0
1,00 1,00 163 49 10,8
0,83 0,67 59 34 12,3
0,83 0,67 187 46 11,3
0,50 0,00 109 55 11,8
0,83 0,00 90 42 7,9
0,17 0,00 105 50 12,7
0,83 1,00 83 13 12,3
1,00 0,67 116 37 11,6
1,00 0,00 42 25 6,7
0,67 0,67 148 30 10,9
1,00 0,00 155 28 12,1
1,00 0,00 125 45 13,3
1,00 0,67 116 35 10,1
0,83 1,00 128 28 5,7
0,33 0,00 138 41 14,3
0,33 0,33 49 6 8,0
1,00 0,67 96 45 13,3
1,00 0,67 164 73 9,3
0,83 0,00 162 17 12,5
1,00 1,00 99 40 7,6
0,83 0,67 202 64 15,9
0,67 0,00 186 37 9,2
0,83 1,00 66 25 9,1
0,67 0,67 183 65 11,1
0,83 0,67 214 100 13,0
0,67 1,00 188 28 14,5
1,00 0,00 104 35 12,2
0,83 0,33 177 56 12,3
0,67 0,67 126 29 11,4
0,83 0,33 76 43 8,8
0,83 0,67 99 59 14,6
0,67 0,00 139 50 12,6
1,00 1,00 78 3 NA
0,33 0,00 162 59 13,0
0,83 0,67 108 27 12,6
1,00 0,33 159 61 13,2
0,83 0,00 74 28 9,9
0,83 0,00 110 51 7,7
0,50 0,33 96 35 10,5
0,50 0,00 116 29 13,4
0,83 0,67 87 48 10,9
1,00 0,33 97 25 4,3
0,33 0,67 127 44 10,3
1,00 0,33 106 64 11,8
0,67 0,33 80 32 11,2
0,83 1,00 74 20 11,4
1,00 0,67 91 28 8,6
0,83 0,00 133 34 13,2
0,83 0,67 74 31 12,6
1,00 0,33 114 26 5,6
0,83 0,00 140 58 9,9
1,00 0,33 95 23 8,8
0,83 0,00 98 21 7,7
0,67 0,00 121 21 9,0
0,83 0,33 126 33 7,3
0,83 0,67 98 16 11,4
0,83 0,67 95 20 13,6
0,83 0,67 110 37 7,9
0,67 0,67 70 35 10,7
0,83 1,00 102 33 10,3
0,00 0,00 86 27 8,3
0,83 0,00 130 41 9,6
1,00 0,00 96 40 14,2
0,17 0,00 102 35 8,5
0,17 0,00 100 28 13,5
0,50 1,00 94 32 4,9
0,50 0,67 52 22 6,4
1,00 0,00 98 44 9,6
0,67 0,67 118 27 11,6
0,83 0,67 99 17 11,1 |