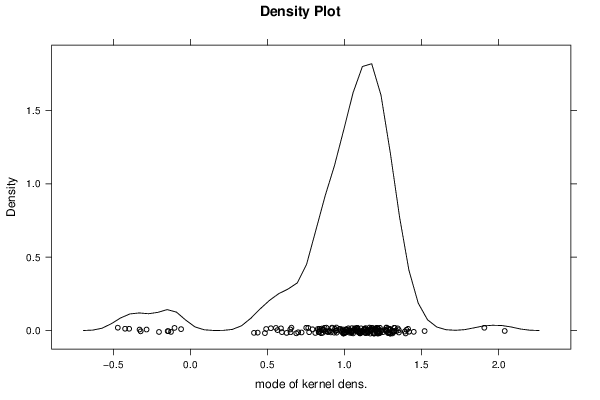
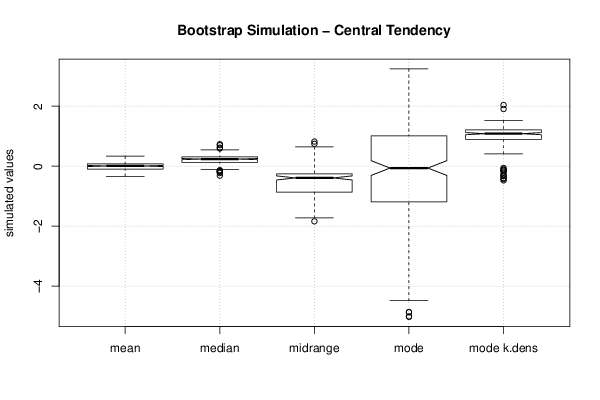
0,943151
1,93488
1,59365
-3,74507
-3,54577
0,74195
3,6163
1,52934
-0,74687
-3,1207
0,701791
-4,73673
1,46245
-1,0326
-1,71871
1,38426
-0,0217053
-0,962395
-2,13819
0,654446
-2,79326
1,976
-0,919187
2,05947
0,714447
-1,45962
1,38903
2,47688
-0,842418
1,25478
-1,22665
0,886873
-0,498445
-3,00844
0,928978
0,470671
0,289022
2,07041
-2,61005
-0,0394285
-4,1215
0,170541
1,5853
-0,029606
2,53209
-0,858974
1,83879
-3,02296
3,31235
2,32646
1,04938
-2,33603
1,24398
1,83566
2,69108
0,26036
-4,86749
4,61513
-0,763781
2,21903
-2,69378
1,9624
-1,52117
3,52463
-1,80004
-0,661775
-1,19354
-0,0449652
3,15832
0,988177
-0,77248
1,15138
-1,72964
2,408
1,85634
1,56628
1,47572
1,79037
-0,380415
-2,44049
-0,589581
3,12272
0,00613861
-5,01623
-1,21834
0,120117
0,922991
1,29604
-2,04363
2,37932
3,05907
-4,48424
-0,78483
-1,32035
-1,9986
-0,954419
-3,3465
1,88184
3,89579
-2,91334
1,14976
0,248063
-1,1974
-1,14895
3,83839
-2,1744
3,24349
-5,00046
-2,78981
-1,1591
1,18015
1,56062
-5,43794
2,17748
2,27391
1,99363
-1,73981
0,592852
-0,826756
1,57612
-3,02344
1,84635
1,11307
5,56521
-2,76439
-0,611745
0,507194
-0,516471
1,62592
0,449954
0,832164
-1,33267
1,00375
1,07947
0,32981
0,384627
3,20101
-0,998316
1,14183
0,772498
-0,677209
2,50498
-0,246527
2,22841
-0,681449
-0,206509
-0,328454
-0,401173
-1,58538
2,03523
-6,0682
0,700179
2,00912
-0,436312
2,301
-0,206327
1,03738
0,970084
-2,01016
-1,9257
1,44366
2,63928
-0,786074
6,51022
0,691773
1,25873
0,227441
1,10766
-0,41337
-2,36348
0,911036
2,26237
-2,85661
-0,885784
-0,855254
2,3495
-1,60338
2,11487
-1,97413
-2,38963
-1,58713
-2,09133
2,00306
-0,300121
-3,73535
0,0611127
2,60331
-2,082
-1,74623
1,66569
-1,16477
0,863274
-2,848
3,18724
-0,169924
1,26748
-2,63909
0,999662
2,24099
0,817941
1,79461
0,519313
-2,58109
0,253274
-1,51395
1,42435
-0,139311
0,987106
1,3655
1,53393
0,0410347
-0,500312
-2,69804
0,39202
-1,18175
-0,885771
0,765205
1,86767
-0,444265
-0,669942
2,12911
-2,7079
-1,33812
0,833997
2,57231
4,12007
1,4694
2,92558
3,07882
2,5567
3,33401
0,826113
0,460824
-2,75847
-1,85633
-5,2214
1,08458
-1,92419
-0,741007
0,165304
-1,66287
-1,47826
0,33342
1,01262
-1,73816
-0,644194
-0,214059
0,643578
-0,654921
-1,88941
-3,12884
-1,02335
1,19346
1,39985
-2,91341
0,564083
-4,23401
-4,00464
-2,83711
-7,29115
0,274851
-1,3576
-2,36969
0,441311
1,34874
0,20435
0,213599
1,35418
1,58484
0,024722
1,16102
-0,238338
1,66962
1,23533
-0,159636
0,00736388
1,06962
-2,19884
|