Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_smp.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
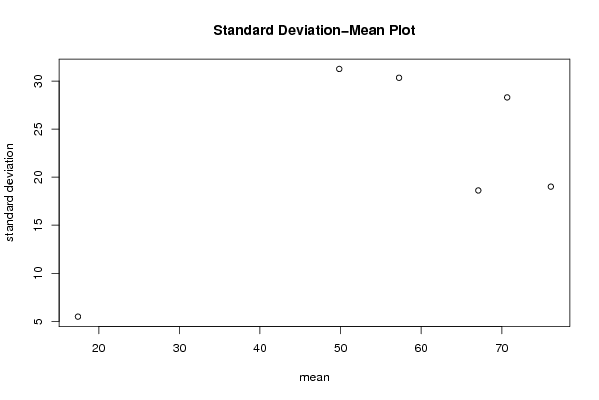
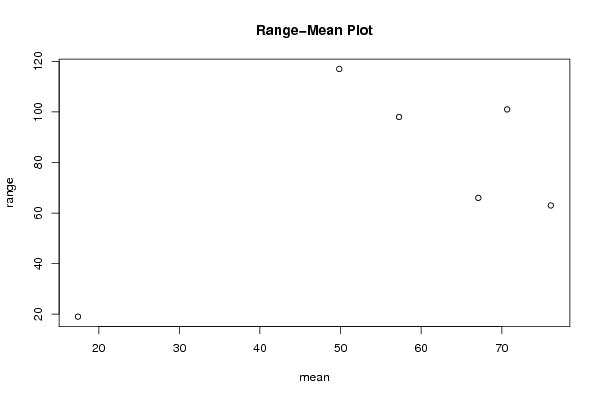
| Title produced by software | Standard Deviation-Mean Plot | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 29 Nov 2010 11:02:42 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2010/Nov/29/t1291028445m7xuo9au9zen2sg.htm/, Retrieved Sat, 27 Apr 2024 06:15:56 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=102829, Retrieved Sat, 27 Apr 2024 06:15:56 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 490 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Univariate Explorative Data Analysis] [Monthly US soldie...] [2010-11-02 12:07:39] [b98453cac15ba1066b407e146608df68] - RMP [Variance Reduction Matrix] [Soldiers] [2010-11-29 09:51:25] [b98453cac15ba1066b407e146608df68] - RM [Standard Deviation-Mean Plot] [Soldiers] [2010-11-29 11:02:42] [d76b387543b13b5e3afd8ff9e5fdc89f] [Current] - RMP [ARIMA Backward Selection] [Soldiers] [2010-11-29 17:56:11] [b98453cac15ba1066b407e146608df68] - PD [ARIMA Backward Selection] [Aantal faillissem...] [2010-12-04 15:51:53] [d6a5e6c1b0014d57cedb2bdfb4a7099f] - [ARIMA Backward Selection] [] [2010-12-07 22:36:55] [afdb2fc47981b6a655b732edc8065db9] - PD [ARIMA Backward Selection] [Verbetering Peer] [2010-12-10 20:50:43] [d6a5e6c1b0014d57cedb2bdfb4a7099f] - R PD [ARIMA Backward Selection] [WS9 4 Foutmelding] [2010-12-07 15:26:08] [afe9379cca749d06b3d6872e02cc47ed] - P [ARIMA Backward Selection] [WS9 4 AR MA] [2010-12-07 15:33:10] [afe9379cca749d06b3d6872e02cc47ed] - D [ARIMA Backward Selection] [] [2010-12-16 15:02:15] [69c775ce4d55db2aa75a88e773e8d700] - R PD [ARIMA Backward Selection] [] [2011-12-03 13:58:43] [74be16979710d4c4e7c6647856088456] - P [ARIMA Backward Selection] [Paper: ARMA] [2011-12-17 16:17:34] [54b1f171ce7a12209ffa11b565e1dcf5] - PD [ARIMA Backward Selection] [Paper: ARMA] [2011-12-17 16:36:06] [54b1f171ce7a12209ffa11b565e1dcf5] - R PD [ARIMA Backward Selection] [ARIMA] [2011-12-04 14:15:32] [74be16979710d4c4e7c6647856088456] - R P [ARIMA Backward Selection] [Workshop 9 - ARIMA] [2011-12-06 18:06:12] [f0855b2dc4da686ca3c0ae4fedd71fda] - P [ARIMA Backward Selection] [] [2012-12-04 03:05:25] [74be16979710d4c4e7c6647856088456] - R P [ARIMA Backward Selection] [WS9 ARIMA Backwar...] [2012-11-29 14:42:51] [617a576b3e2f0c57f6da5ea5fef54049] - P [ARIMA Backward Selection] [Arima backward se...] [2012-12-07 09:43:48] [57fcaae991493f873bcb4ee93ca06ef0] - R PD [ARIMA Backward Selection] [WS 9 ARIMA Backwa...] [2012-12-03 19:03:37] [8c30f4dd45e15fd207e4faf2fdf6253e] - R P [ARIMA Backward Selection] [] [2012-12-04 16:26:05] [74be16979710d4c4e7c6647856088456] - R P [ARIMA Backward Selection] [workshop 9: AR MA...] [2012-12-04 18:23:48] [74be16979710d4c4e7c6647856088456] - RMP [ARIMA Backward Selection] [WS 9 - ARIMA] [2012-12-04 18:58:07] [74be16979710d4c4e7c6647856088456] - D [ARIMA Backward Selection] [WS9] [2010-12-07 21:01:45] [87116ee6ef949037dfa02b8eb1a3bf97] - PD [ARIMA Backward Selection] [Correctie WS9] [2010-12-13 20:14:37] [87116ee6ef949037dfa02b8eb1a3bf97] - PD [ARIMA Backward Selection] [ARIMA ] [2010-12-15 12:36:54] [0ed8ad64bdfc801eaa95d5097964fc04] - [ARIMA Backward Selection] [Workshop 9 (ARIMA)] [2010-12-15 16:00:56] [845827b7f02503df17c96f445745fee7] - [ARIMA Backward Selection] [] [2010-12-16 16:59:52] [24bb5b06bd1854f48aebec8f44957ed0] - P [ARIMA Backward Selection] [ARIMA Backward Se...] [2010-12-16 14:05:46] [9f313cc7203314d73bf17d2b325aee79] - RMP [ARIMA Forecasting] [ARIMA Forecasting] [2010-12-16 14:17:27] [9f313cc7203314d73bf17d2b325aee79] - PD [ARIMA Backward Selection] [estimating arma p...] [2010-12-22 16:03:39] [a8a0ff0853b70f438be515083758c362] - PD [ARIMA Backward Selection] [WS 9: ARIMA backw...] [2011-12-01 13:47:14] [10b12745961ee885a66356b3bf31ed40] - [ARIMA Backward Selection] [WS 9 - ARIMA ] [2011-12-02 13:57:49] [6a3e51c0c7ab195427042dfaef1df5a0] - PD [ARIMA Backward Selection] [Model 1] [2011-12-05 11:47:38] [c035d973aa8488be257660c2dc4ec375] - R D [ARIMA Backward Selection] [Workshop 9 (5)] [2011-12-06 09:02:52] [74be16979710d4c4e7c6647856088456] - PD [ARIMA Backward Selection] [Workshop 9: ARIMA...] [2011-12-06 09:56:00] [21b3d52ef28595defb5676e0f3570994] - R PD [ARIMA Backward Selection] [WS IX-aantal over...] [2011-12-06 20:15:53] [74be16979710d4c4e7c6647856088456] - P [ARIMA Backward Selection] [WS IX-aantal over...] [2011-12-06 20:44:23] [74be16979710d4c4e7c6647856088456] - D [ARIMA Backward Selection] [Paper arima foutm...] [2011-12-20 16:15:18] [7c680a04865e75aa8ab422cdbfd97ac3] - P [ARIMA Backward Selection] [Paper arima corre...] [2011-12-22 11:17:52] [7c680a04865e75aa8ab422cdbfd97ac3] - RMPD [Variance Reduction Matrix] [Variance reductio...] [2011-12-07 09:43:44] [71fa56397b79b6f4596b21687a2f6e82] - RMPD [(Partial) Autocorrelation Function] [ACF d=0 en D=1] [2011-12-07 09:58:09] [71fa56397b79b6f4596b21687a2f6e82] - RMP [Spectral Analysis] [spectraalanalyse] [2011-12-08 21:23:11] [71fa56397b79b6f4596b21687a2f6e82] - RMPD [(Partial) Autocorrelation Function] [ACF d=0 en D=0] [2011-12-07 10:03:24] [71fa56397b79b6f4596b21687a2f6e82] - RMPD [(Partial) Autocorrelation Function] [ACF d=1 en D=0] [2011-12-07 10:04:06] [71fa56397b79b6f4596b21687a2f6e82] - RMPD [(Partial) Autocorrelation Function] [ACF d=1 en D=1] [2011-12-07 10:04:53] [71fa56397b79b6f4596b21687a2f6e82] - D [ARIMA Backward Selection] [ARIMA Backward Se...] [2011-12-07 12:35:23] [933c43040ae019b88c07d6cc7db3263c] - RMPD [(Partial) Autocorrelation Function] [] [2011-12-07 18:04:11] [5b1044653d12da6c563533920760fdbb] - RMPD [(Partial) Autocorrelation Function] [] [2011-12-07 18:10:28] [5b1044653d12da6c563533920760fdbb] - RMP [Stem-and-leaf Plot] [] [2011-12-07 18:11:06] [5b1044653d12da6c563533920760fdbb] - RMP [Spectral Analysis] [] [2011-12-07 18:16:08] [5b1044653d12da6c563533920760fdbb] - RMP [Variance Reduction Matrix] [] [2011-12-07 18:18:13] [5b1044653d12da6c563533920760fdbb] [Truncated] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
37 30 47 35 30 43 82 40 47 19 52 136 80 42 54 66 81 63 137 72 107 58 36 52 79 77 54 84 48 96 83 66 61 53 30 74 69 59 42 65 70 100 63 105 82 81 75 102 121 98 76 77 63 37 35 23 40 29 37 51 20 28 13 22 25 13 16 13 16 17 9 17 25 14 8 7 10 7 10 3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = 0 ; par3 = 0 ; par4 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||