Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_fitdistrnorm.wasp | ||||||||||||||||||||||||||||||||||||||
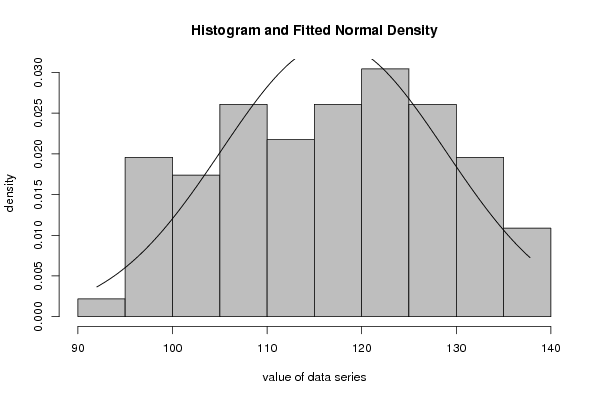
| Title produced by software | Maximum-likelihood Fitting - Normal Distribution | ||||||||||||||||||||||||||||||||||||||
| Date of computation | Wed, 12 Nov 2008 06:54:54 -0700 | ||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2008/Nov/12/t1226498128kqeac7ei3cp4253.htm/, Retrieved Sun, 19 May 2024 05:48:19 +0000 | ||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=24189, Retrieved Sun, 19 May 2024 05:48:19 +0000 | |||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 173 | ||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||
| F [Univariate Data Series] [Niet werkende wer...] [2008-10-13 17:30:07] [91d2608132ba5d00ecce3524d8276757] F RMPD [Partial Correlation] [Partial Correlati...] [2008-11-11 19:49:25] [deb3c14ac9e4607a6d84fc9d0e0e6cc2] F D [Partial Correlation] [Partial Correlati...] [2008-11-11 19:56:04] [deb3c14ac9e4607a6d84fc9d0e0e6cc2] F RM D [Box-Cox Linearity Plot] [Box-Cox Linearity...] [2008-11-11 21:16:20] [deb3c14ac9e4607a6d84fc9d0e0e6cc2] F RM D [Box-Cox Normality Plot] [Box-Cox Normality...] [2008-11-11 21:26:55] [deb3c14ac9e4607a6d84fc9d0e0e6cc2] F RM [Maximum-likelihood Fitting - Normal Distribution] [Maximum-likelihoo...] [2008-11-11 21:28:51] [deb3c14ac9e4607a6d84fc9d0e0e6cc2] F PD [Maximum-likelihood Fitting - Normal Distribution] [] [2008-11-12 13:54:54] [890f958132adb51f538eb267509d2e70] [Current] | |||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||
98.6 98 106.8 96.7 100.2 107.7 92 98.4 107.4 117.7 105.7 97.5 99.9 98.2 104.5 100.8 101.5 103.9 99.6 98.4 112.7 118.4 108.1 105.4 114.6 106.9 115.9 109.8 101.8 114.2 110.8 108.4 127.5 128.6 116.6 127.4 105 108.3 125 111.6 106.5 130.3 115 116.1 134 126.5 125.8 136.4 114.9 110.9 125.5 116.8 116.8 125.5 104.2 115.1 132.8 123.3 124.8 122 117.4 117.9 137.4 114.6 124.7 129.6 109.4 120.9 134.9 136.3 133.2 127.2 122.7 120.5 137.8 119.1 124.3 134.4 121.1 122.2 127.7 137.4 132.2 129.2 124.9 124.8 128.2 134.4 118.6 132.6 123.2 112.3 | |||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||
| par1 = 8 ; par2 = 0 ; | |||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||
| par1 = 8 ; par2 = 0 ; | |||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||
library(MASS) | |||||||||||||||||||||||||||||||||||||||