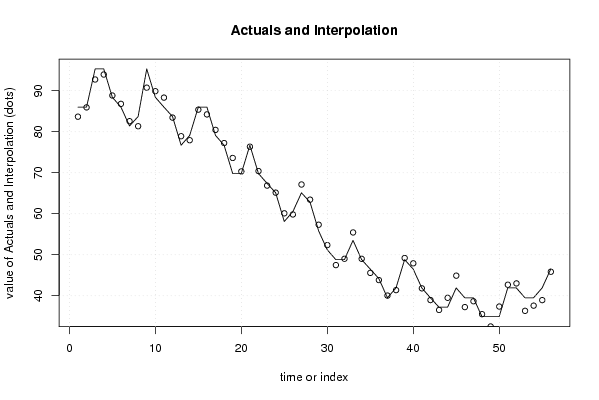
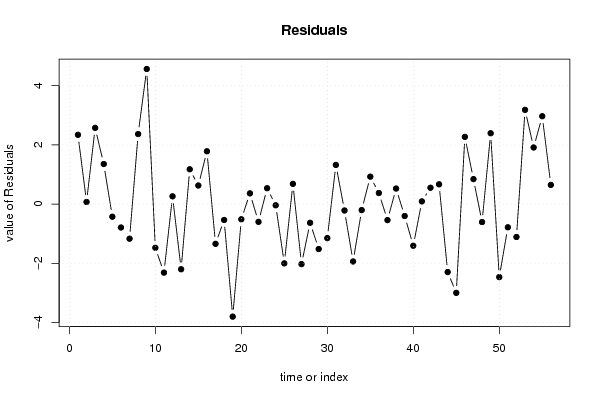
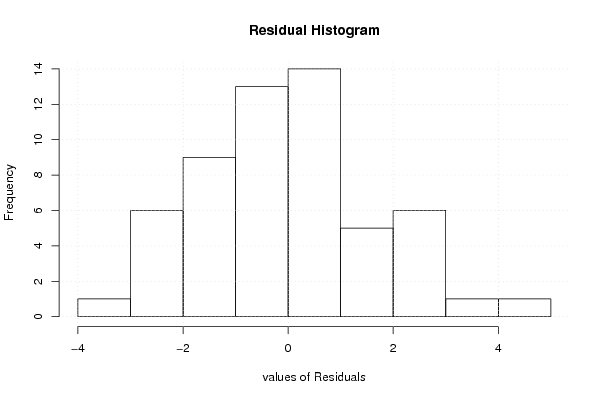
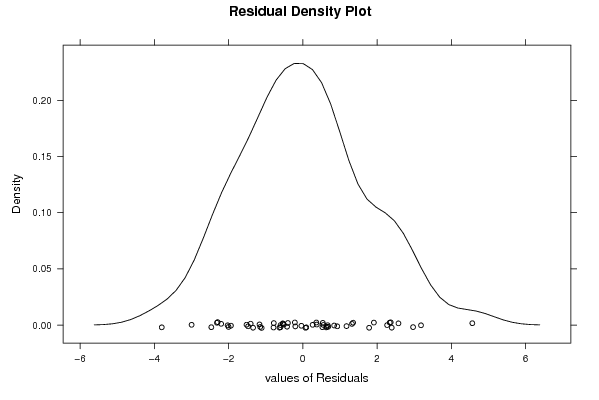
86,0 88,4 90,7 95,3 100,0 94,7
86,0 86,0 88,4 90,7 95,3 110,6
95,3 86,0 86,0 88,4 90,7 71,3
95,3 95,3 86,0 86,0 88,4 104,1
88,4 95,3 95,3 86,0 86,0 112,3
86,0 88,4 95,3 95,3 86,0 110,2
81,4 86,0 88,4 95,3 95,3 112,9
83,7 81,4 86,0 88,4 95,3 95,1
95,3 83,7 81,4 86,0 88,4 103,1
88,4 95,3 83,7 81,4 86,0 101,9
86,0 88,4 95,3 83,7 81,4 100,4
83,7 86,0 88,4 95,3 83,7 106,9
76,7 83,7 86,0 88,4 95,3 100,7
79,1 76,7 83,7 86,0 88,4 114,3
86,0 79,1 76,7 83,7 86,0 73,3
86,0 86,0 79,1 76,7 83,7 105,9
79,1 86,0 86,0 79,1 76,7 113,9
76,7 79,1 86,0 86,0 79,1 112,1
69,8 76,7 79,1 86,0 86,0 117,5
69,8 69,8 76,7 79,1 86,0 97,5
76,7 69,8 69,8 76,7 79,1 112,3
69,8 76,7 69,8 69,8 76,7 106,9
67,4 69,8 76,7 69,8 69,8 120,9
65,1 67,4 69,8 76,7 69,8 92,7
58,1 65,1 67,4 69,8 76,7 110,9
60,5 58,1 65,1 67,4 69,8 116,5
65,1 60,5 58,1 65,1 67,4 77,1
62,8 65,1 60,5 58,1 65,1 113,1
55,8 62,8 65,1 60,5 58,1 115,9
51,2 55,8 62,8 65,1 60,5 123,5
48,8 51,2 55,8 62,8 65,1 123,6
48,8 48,8 51,2 55,8 62,8 101,5
53,5 48,8 48,8 51,2 55,8 121,0
48,8 53,5 48,8 48,8 51,2 112,2
46,5 48,8 53,5 48,8 48,8 126,0
44,2 46,5 48,8 53,5 48,8 101,8
39,5 44,2 46,5 48,8 53,5 117,9
41,9 39,5 44,2 46,5 48,8 122,2
48,8 41,9 39,5 44,2 46,5 82,7
46,5 48,8 41,9 39,5 44,2 120,5
41,9 46,5 48,8 41,9 39,5 120,3
39,5 41,9 46,5 48,8 41,9 134,2
37,2 39,5 41,9 46,5 48,8 128,2
37,2 37,2 39,5 41,9 46,5 100,5
41,9 37,2 37,2 39,5 41,9 126,0
39,5 41,9 37,2 37,2 39,5 122,9
39,5 39,5 41,9 37,2 37,2 106,1
34,9 39,5 39,5 41,9 37,2 130,4
34,9 34,9 39,5 39,5 41,9 121,3
34,9 34,9 34,9 39,5 39,5 126,1
41,9 34,9 34,9 34,9 39,5 88,7
41,9 41,9 34,9 34,9 34,9 118,7
39,5 41,9 41,9 34,9 34,9 129,3
39,5 39,5 41,9 41,9 34,9 136,2
41,9 39,5 39,5 41,9 41,9 123,0
46,5 41,9 39,5 39,5 41,9 103,5
|