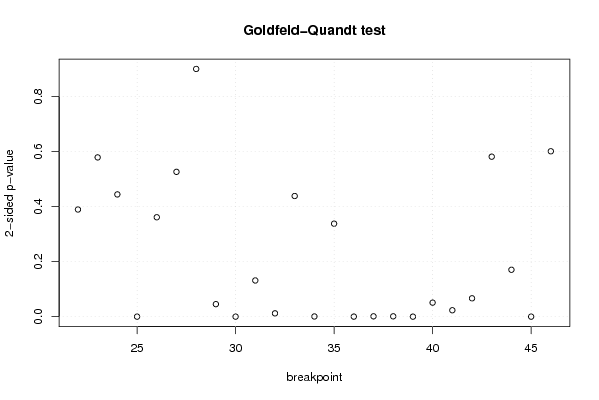
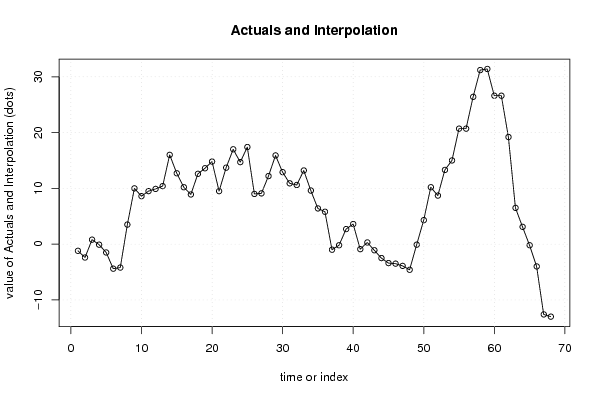
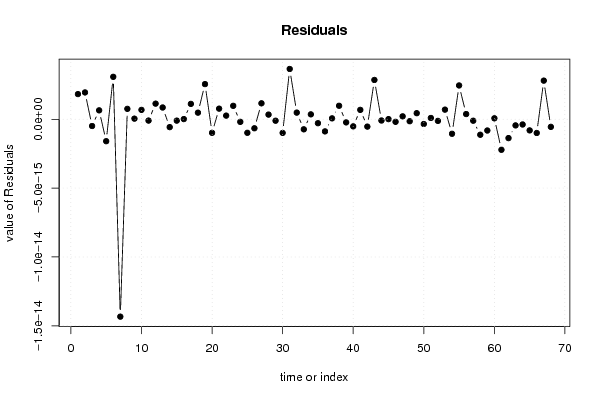
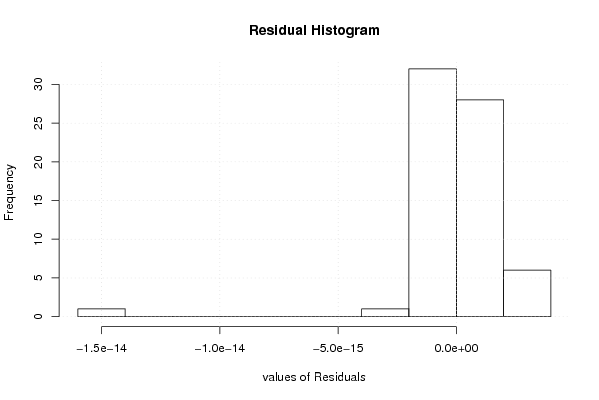
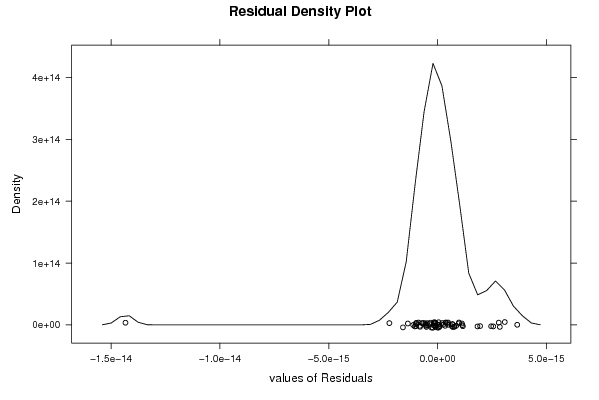
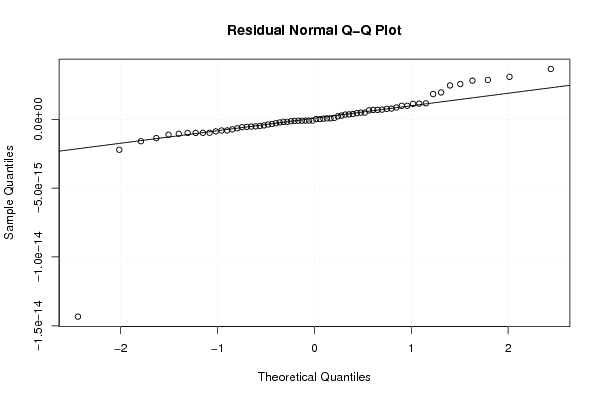
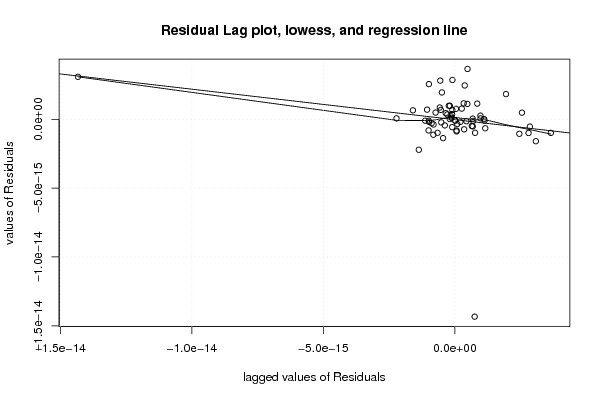
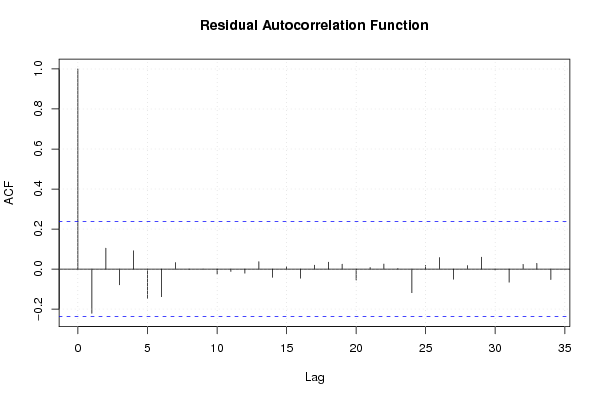
-1,2 23,6 -1,5 -0,1 0,8 -2,4 -1,2
-2,4 25,7 -4,4 -1,5 -0,1 0,8 -2,4
0,8 32,5 -4,2 -4,4 -1,5 -0,1 0,8
-0,1 33,5 3,5 -4,2 -4,4 -1,5 -0,1
-1,5 34,5 10 3,5 -4,2 -4,4 -1,5
-4,4 27,9 8,6 10 3,5 -4,2 -4,4
-4,2 45,3 9,5 8,6 10 3,5 -4,2
3,5 40,8 9,9 9,5 8,6 10 3,5
10 58,5 10,4 9,9 9,5 8,6 10
8,6 32,5 16 10,4 9,9 9,5 8,6
9,5 35,5 12,7 16 10,4 9,9 9,5
9,9 46,7 10,2 12,7 16 10,4 9,9
10,4 53,2 8,9 10,2 12,7 16 10,4
16 36,1 12,6 8,9 10,2 12,7 16
12,7 54 13,6 12,6 8,9 10,2 12,7
10,2 58,1 14,8 13,6 12,6 8,9 10,2
8,9 41,8 9,5 14,8 13,6 12,6 8,9
12,6 43,1 13,7 9,5 14,8 13,6 12,6
13,6 76 17 13,7 9,5 14,8 13,6
14,8 42,8 14,7 17 13,7 9,5 14,8
9,5 41 17,4 14,7 17 13,7 9,5
13,7 61,4 9 17,4 14,7 17 13,7
17 34,2 9,1 9 17,4 14,7 17
14,7 53,8 12,2 9,1 9 17,4 14,7
17,4 80,7 15,9 12,2 9,1 9 17,4
9 79,5 12,9 15,9 12,2 9,1 9
9,1 96,5 10,9 12,9 15,9 12,2 9,1
12,2 108,3 10,6 10,9 12,9 15,9 12,2
15,9 100,1 13,2 10,6 10,9 12,9 15,9
12,9 108,5 9,6 13,2 10,6 10,9 12,9
10,9 127,4 6,4 9,6 13,2 10,6 10,9
10,6 86,5 5,8 6,4 9,6 13,2 10,6
13,2 71,4 -1 5,8 6,4 9,6 13,2
9,6 88,2 -0,2 -1 5,8 6,4 9,6
6,4 135,6 2,7 -0,2 -1 5,8 6,4
5,8 70,5 3,6 2,7 -0,2 -1 5,8
-1 87,5 -0,9 3,6 2,7 -0,2 -1
-0,2 73,3 0,3 -0,9 3,6 2,7 -0,2
2,7 92,2 -1,1 0,3 -0,9 3,6 2,7
3,6 61,1 -2,5 -1,1 0,3 -0,9 3,6
-0,9 45,7 -3,4 -2,5 -1,1 0,3 -0,9
0,3 30,5 -3,5 -3,4 -2,5 -1,1 0,3
-1,1 34,8 -3,9 -3,5 -3,4 -2,5 -1,1
-2,5 29,2 -4,6 -3,9 -3,5 -3,4 -2,5
-3,4 56,7 -0,1 -4,6 -3,9 -3,5 -3,4
-3,5 67,1 4,3 -0,1 -4,6 -3,9 -3,5
-3,9 41,8 10,2 4,3 -0,1 -4,6 -3,9
-4,6 46,8 8,7 10,2 4,3 -0,1 -4,6
-0,1 50,1 13,3 8,7 10,2 4,3 -0,1
4,3 81,9 15 13,3 8,7 10,2 4,3
10,2 115,8 20,7 15 13,3 8,7 10,2
8,7 102,5 20,7 20,7 15 13,3 8,7
13,3 106,6 26,4 20,7 20,7 15 13,3
15 101,4 31,2 26,4 20,7 20,7 15
20,7 136,1 31,4 31,2 26,4 20,7 20,7
20,7 143,4 26,6 31,4 31,2 26,4 20,7
26,4 127,5 26,6 26,6 31,4 31,2 26,4
31,2 113,8 19,2 26,6 26,6 31,4 31,2
31,4 75,3 6,5 19,2 26,6 26,6 31,4
26,6 98,5 3,1 6,5 19,2 26,6 26,6
26,6 113,7 -0,2 3,1 6,5 19,2 26,6
19,2 103,7 -4 -0,2 3,1 6,5 19,2
6,5 73,9 -12,6 -4 -0,2 3,1 6,5
3,1 52,5 -13 -12,6 -4 -0,2 3,1
-0,2 63,9 -17,6 -13 -12,6 -4 -0,2
-4 44,9 -21,7 -17,6 -13 -12,6 -4
-12,6 31,3 -23,2 -21,7 -17,6 -13 -12,6
-13 24,9 -16,8 -23,2 -21,7 -17,6 -13
|