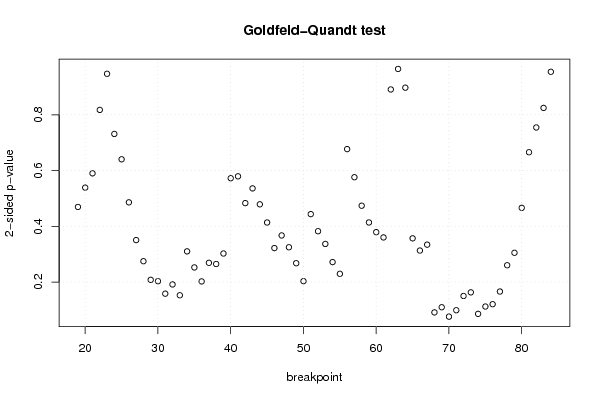
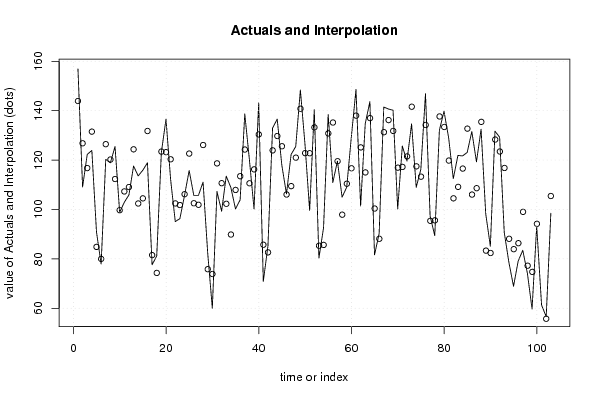
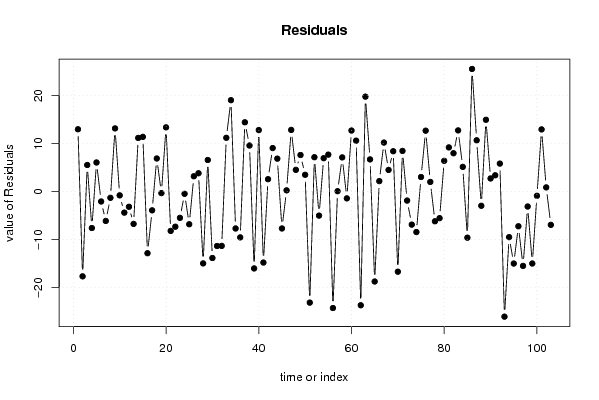
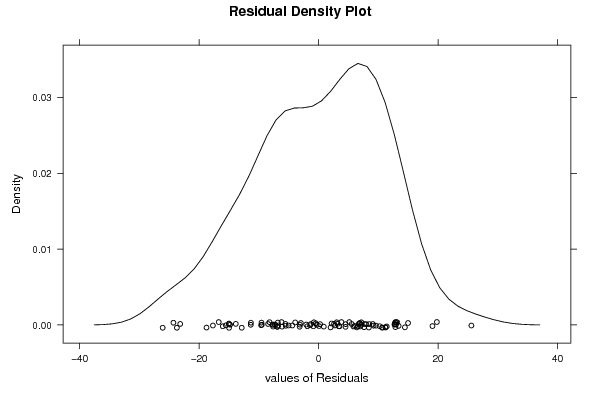
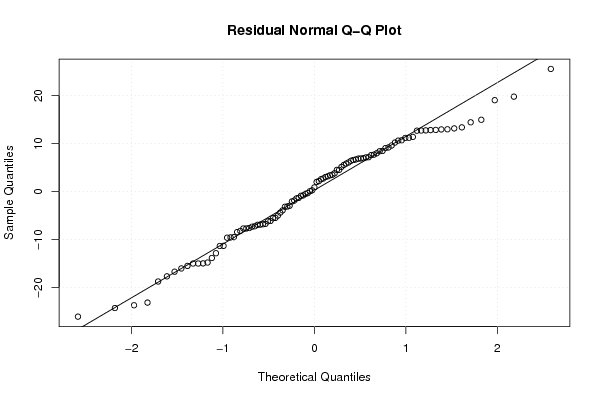
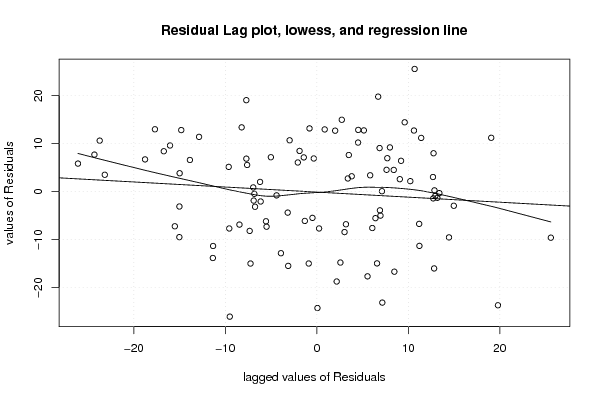
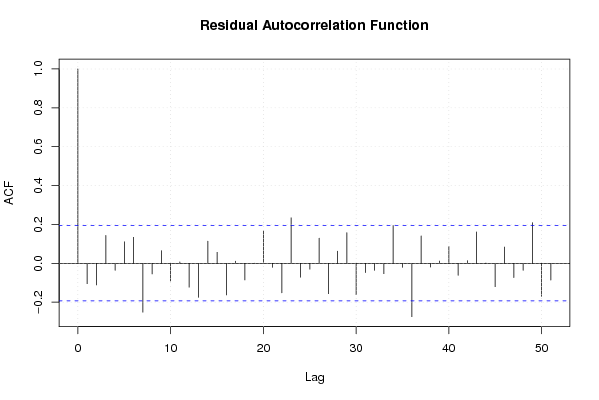
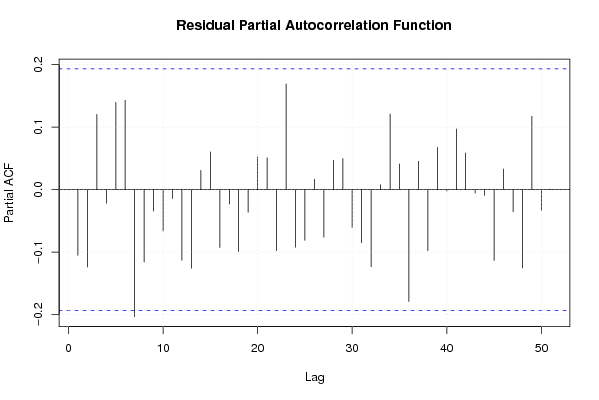
156,9 0 136,9 128,7
109,1 0 156,9 136,9
122,3 0 109,1 156,9
123,9 0 122,3 109,1
90,9 0 123,9 122,3
77,9 0 90,9 123,9
120,3 0 77,9 90,9
118,9 0 120,3 77,9
125,5 0 118,9 120,3
98,9 0 125,5 118,9
102,9 0 98,9 125,5
105,9 0 102,9 98,9
117,6 0 105,9 102,9
113,6 0 117,6 105,9
115,9 0 113,6 117,6
118,9 0 115,9 113,6
77,6 0 118,9 115,9
81,2 0 77,6 118,9
123,1 0 81,2 77,6
136,6 0 123,1 81,2
112,1 0 136,6 123,1
95,1 0 112,1 136,6
96,3 0 95,1 112,1
105,7 0 96,3 95,1
115,8 0 105,7 96,3
105,7 0 115,8 105,7
105,7 0 105,7 115,8
111,1 0 105,7 105,7
82,4 0 111,1 105,7
60 0 82,4 111,1
107,3 0 60 82,4
99,3 0 107,3 60
113,5 0 99,3 107,3
108,9 0 113,5 99,3
100,2 0 108,9 113,5
103,9 0 100,2 108,9
138,7 0 103,9 100,2
120,2 0 138,7 103,9
100,2 0 120,2 138,7
143,2 0 100,2 120,2
70,9 0 143,2 100,2
85,2 0 70,9 143,2
133 0 85,2 70,9
136,6 0 133 85,2
117,9 0 136,6 133
106,3 0 117,9 136,6
122,3 0 106,3 117,9
125,5 0 122,3 106,3
148,4 0 125,5 122,3
126,3 0 148,4 125,5
99,6 0 126,3 148,4
140,4 0 99,6 126,3
80,3 0 140,4 99,6
92,6 0 80,3 140,4
138,5 0 92,6 80,3
110,9 0 138,5 92,6
119,6 0 110,9 138,5
105 0 119,6 110,9
109 0 105 119,6
129,4 0 109 105
148,6 0 129,4 109
101,4 0 148,6 129,4
134,8 0 101,4 148,6
143,7 0 134,8 101,4
81,6 0 143,7 134,8
90,3 0 81,6 143,7
141,5 0 90,3 81,6
140,7 0 141,5 90,3
140,2 0 140,7 141,5
100,2 0 140,2 140,7
125,7 0 100,2 140,2
119,6 0 125,7 100,2
134,7 0 119,6 125,7
109 0 134,7 119,6
116,3 0 109 134,7
146,9 0 116,3 109
97,4 0 146,9 116,3
89,4 0 97,4 146,9
132,1 0 89,4 97,4
139,8 0 132,1 89,4
129 1 139,8 132,1
112,5 1 129 139,8
121,9 1 112,5 129
121,7 1 121,9 112,5
123,1 1 121,7 121,9
131,6 1 123,1 121,7
119,3 1 131,6 123,1
132,5 1 119,3 131,6
98,3 1 132,5 119,3
85,1 1 98,3 132,5
131,7 1 85,1 98,3
129,3 1 131,7 85,1
90,7 1 129,3 131,7
78,6 1 90,7 129,3
68,9 1 78,6 90,7
79,1 1 68,9 78,6
83,5 1 79,1 68,9
74,1 1 83,5 79,1
59,7 1 74,1 83,5
93,3 1 59,7 74,1
61,3 1 93,3 59,7
56,6 1 61,3 93,3
98,5 1 56,6 61,3
|