Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_edabi.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Bivariate Explorative Data Analysis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Wed, 04 Nov 2009 04:34:29 -0700 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2009/Nov/04/t1257334595dkyiws25odngdsv.htm/, Retrieved Mon, 29 Apr 2024 12:02:57 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=53566, Retrieved Mon, 29 Apr 2024 12:02:57 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | WS 5 (Et inY en E’t in X) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 184 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Bivariate Data Series] [Bivariate dataset] [2008-01-05 23:51:08] [74be16979710d4c4e7c6647856088456] - RMPD [Bivariate Explorative Data Analysis] [] [2009-10-28 15:16:32] [5482608004c1d7bbf873930172393a2d] - D [Bivariate Explorative Data Analysis] [] [2009-10-28 16:44:36] [5482608004c1d7bbf873930172393a2d] - RMPD [Trivariate Scatterplots] [] [2009-11-03 17:57:36] [5482608004c1d7bbf873930172393a2d] - RMPD [Bivariate Explorative Data Analysis] [] [2009-11-03 18:33:00] [5482608004c1d7bbf873930172393a2d] - D [Bivariate Explorative Data Analysis] [Workshop 5 Bivari...] [2009-11-04 11:34:29] [e2f800c9186517d2e5c4a809848912a7] [Current] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
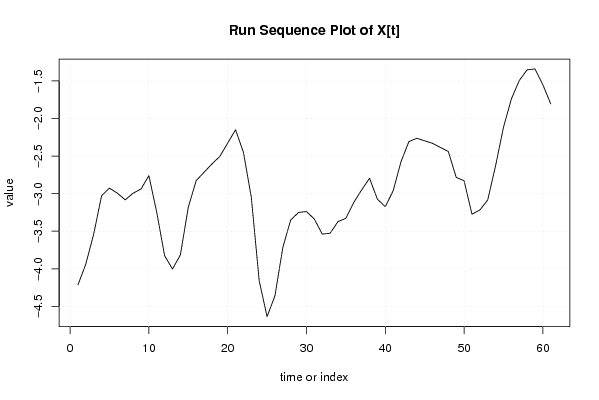
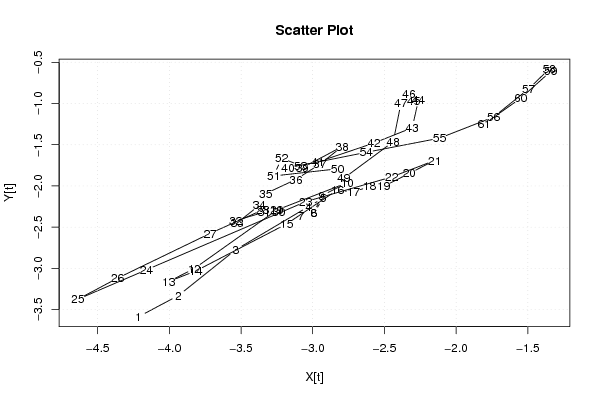
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-4.213500441 -3.935935693 -3.536894327 -3.026635334 -2.926635334 -2.992982456 -3.08176483 -2.992982456 -2.93785296 -2.760288212 -3.249070586 -3.825676701 -4.003241449 -3.814459075 -3.18176483 -2.826635334 -2.715417708 -2.604200082 -2.504200082 -2.326635334 -2.149070586 -2.449070586 -3.049070586 -4.159329578 -4.636894327 -4.359329578 -3.715417708 -3.349070586 -3.249070586 -3.23785296 -3.33785296 -3.53785296 -3.526635334 -3.371505838 -3.327593967 -3.116376341 -2.950029219 -2.794899723 -3.072464471 -3.172464471 -2.961246845 -2.572464471 -2.306117349 -2.262205478 -2.295858356 -2.329511234 -2.38464073 -2.438811593 -2.782723464 -2.827593967 -3.272464471 -3.217334975 -3.083682097 -2.627593967 -2.116376341 -1.738811593 -1.494899723 -1.350987852 -1.339770226 -1.550029219 -1.805158715 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
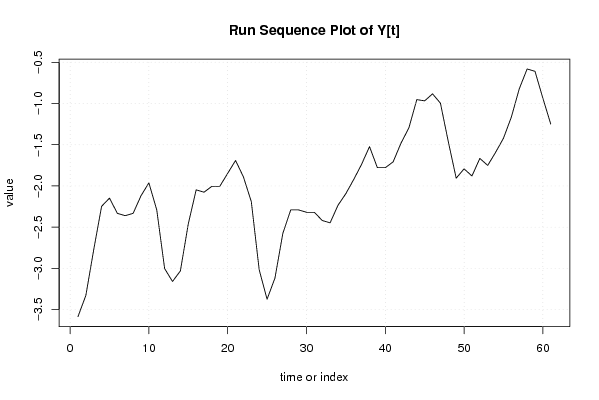
| Dataseries Y: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-3.585860839 -3.329201771 -2.774235967 -2.247599696 -2.147599696 -2.332588297 -2.360917831 -2.332588297 -2.119270162 -1.962611095 -2.290940628 -3.0025655 -3.159224568 -3.030895034 -2.460917831 -2.047599696 -2.07592923 -2.004258764 -2.004258764 -1.847599696 -1.690940628 -1.890940628 -2.190940628 -3.017576899 -3.374235967 -3.117576899 -2.57592923 -2.290940628 -2.290940628 -2.319270162 -2.319270162 -2.419270162 -2.447599696 -2.234281561 -2.092633892 -1.920963426 -1.735974824 -1.522656689 -1.779315756 -1.779315756 -1.70764529 -1.479315756 -1.294327155 -0.952679486 -0.967690884 -0.882702283 -0.996020418 -1.464304358 -1.905952027 -1.792633892 -1.879315756 -1.665997621 -1.750986223 -1.592633892 -1.420963426 -1.164304358 -0.822656689 -0.58100902 -0.609338554 -0.935974824 -1.249292959 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
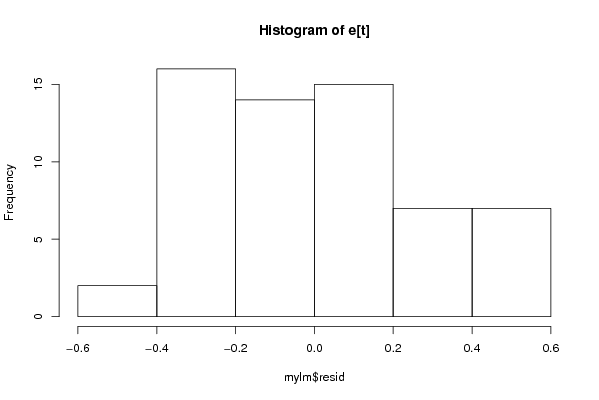
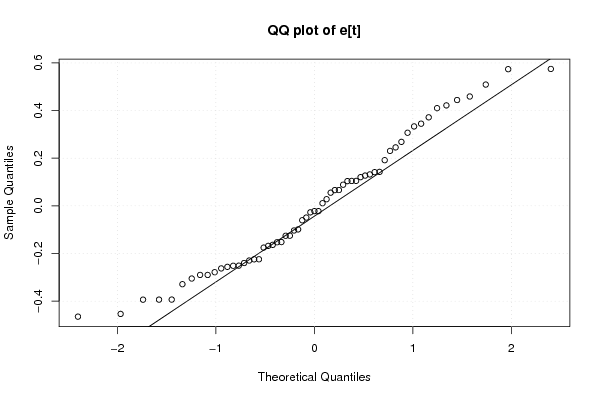
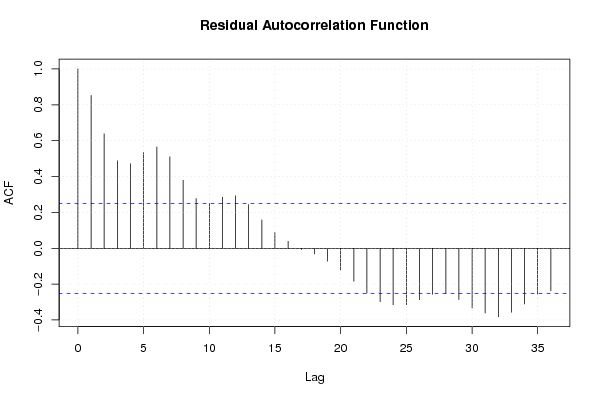
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = 36 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = 36 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||