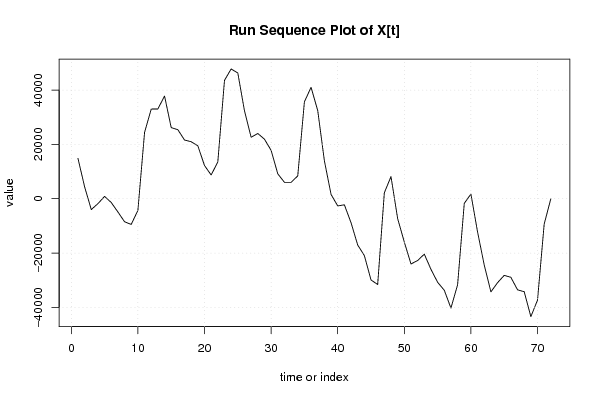
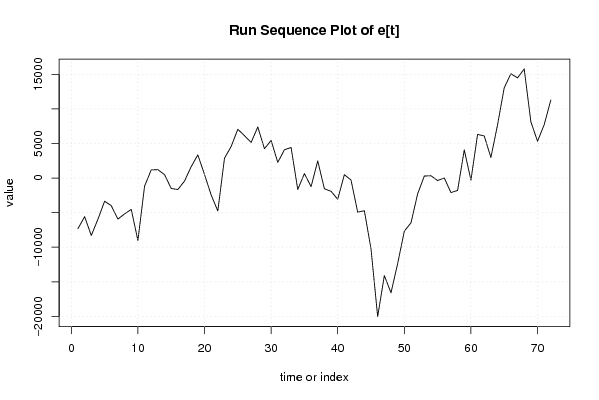
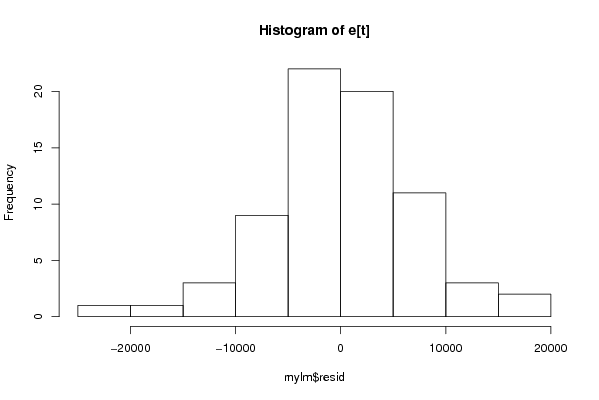
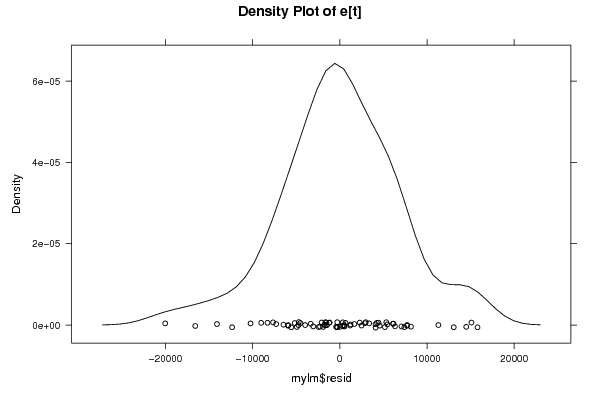
14836,78129
4363,00051
-4002,328314
-1812,109098
848,00051
-1406,670666
-4893,45145
-8497,328314
-9477,095569
-4271,766746
24414,12365
33009,34286
33054,56208
37817,57561
26184,90443
25413,23325
21611,23325
20980,90443
19480,35639
12208,68521
8744,014038
13570,57561
43565,35639
47787,35639
46298,24678
32359,01404
22631,90443
24020,57561
21922,90443
17815,12365
9175,890902
6032,562078
6020,342862
8397,671686
35701,00051
41019,00051
32444,43894
13947,43894
1575,110118
-2650,99949
-2245,109098
-8832,218706
-17035,21871
-20944,21871
-29913,67067
-31604,67067
2188,219726
8149,54855
-7346,889882
-15975,65714
-24040,42439
-22757,64361
-20422,08204
-26004,19165
-30769,06851
-33706,8493
-40292,94538
-31693,60302
-1712,712632
1714,835408
-12289,2742
-24404,39734
-34307,64361
-30919,09557
-28222,76675
-28942,32831
-33556,90341
-34260,90341
-43434,80733
-37319,04008
-9341,382428
3,631100644 |
523,5658454
-3273,146241
-10414,07811
-6902,790198
-2897,146241
-4727,214371
-8504,926458
-9664,078111
-9522,585808
-11281,65394
11711,70211
18606,99002
18669,27793
20458,48232
12320,41419
11765,34606
10978,34606
12736,41419
13642,19441
6972,126279
2179,058149
2409,482322
25864,19441
29791,19441
31499,55045
23225,05815
17110,41419
20072,48232
15823,41419
14852,70211
7120,209802
7283,277932
7607,990019
2783,921889
19491,85376
20415,85376
19612,42959
5818,429585
-1078,502285
-4449,146241
-681,7901979
-4945,434155
-13923,43415
-15768,43415
-26036,21437
-36721,21437
-12942,85833
-12288,92646
-16241,50228
-16101,00998
-19161,51768
-14289,80559
-10476,38142
-13384,02537
-16588,17703
-17771,88911
-23359,32868
-18508,19242
3181,163623
608,3834059
-161,2605507
-6787,108898
-15131,80559
-8601,585808
-1830,653938
-180,0781112
-3207,706675
-2290,706675
-14767,26711
-14364,75941
2786,104328
11306,30872 |