Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_edabi.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Bivariate Explorative Data Analysis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Wed, 04 Nov 2009 13:57:34 -0700 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2009/Nov/04/t1257368364ra5n0wjsuxu6p0t.htm/, Retrieved Mon, 29 Apr 2024 13:26:06 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=53847, Retrieved Mon, 29 Apr 2024 13:26:06 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | ws5bel36mldg | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 103 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Bivariate Explorative Data Analysis] [WS5: Bivariate ED...] [2009-11-04 20:57:34] [3d2053c5f7c50d3c075d87ce0bd87294] [Current] - RMPD [Kendall tau Rank Correlation] [WS 5: Kendall ran...] [2009-11-04 21:11:24] [7c2a5b25a196bd646844b8f5223c9b3e] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
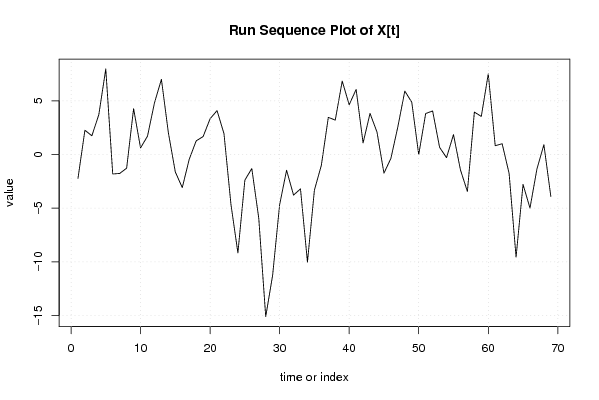
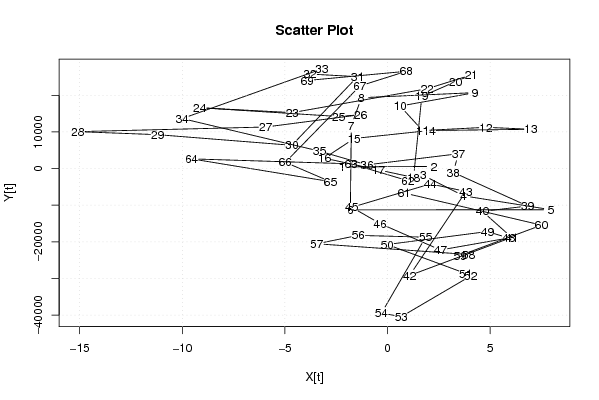
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-2,225348423 2,257563984 1,740476391 3,706301205 7,959396904 -1,810786388 -1,764228405 -1,270419804 4,26994678 0,614671842 1,706301205 4,797930568 6,99357209 2,027747276 -1,627527662 -3,076264883 -0,444268937 1,268460179 1,670639417 3,325914355 4,085201453 1,936117913 -4,638423857 -9,15551145 -2,385328158 -1,30459499 -5,936590936 -15,08567448 -11,20058283 -4,660216246 -1,464574724 -3,7751246 -3,20058283 -10,00494131 -3,315491184 -1,004941308 3,454692108 3,201596409 6,833592356 4,627054639 6,052512869 1,07579186 3,822696161 2,086688055 -1,740949414 -0,362049167 2,580496656 5,901942727 4,867767541 -0,017324105 3,810313364 4,051026267 0,672126019 -0,293698795 1,855384745 -1,43188614 -3,444268937 3,945527506 3,541861665 7,506199877 0,816749754 1,000008479 -1,770174813 -9,549075061 -2,783250246 -4,981417326 -1,341050742 0,916057116 -3,913759591 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
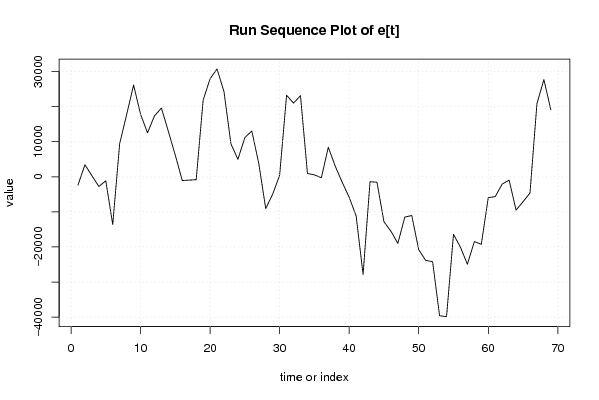
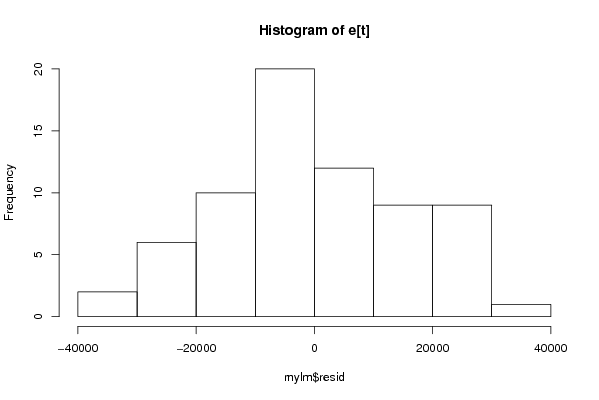
| Dataseries Y: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
453.76 567.15 -1861.46 -7454.68 -11242.01 -11289.29 11715.62 19336.27 20729.84 17062.92 10369.32 11258.72 10721.21 10375.43 8202.51 2834.34 -371.69 -2429.58 19711.67 23745.59 25554.61 21797.03 15266.24 16605.63 14210.92 14714.05 11338.08 10074.50 9246.14 6347.71 25088.21 25798.35 27194.14 13649.64 4755.79 1004.64 4035.07 -1054.60 -10228.63 -11699.39 -18817.18 -29199.23 -6246.90 -4201.96 -10585.42 -15033.13 -22236.31 -18963.19 -17206.41 -20744.05 -28649.59 -29339.61 -40442.91 -39478.68 -18740.10 -18252.00 -20542.69 -23465.13 -23725.81 -15453.46 -6685.61 -3334.80 1292.91 2637.62 -3574.61 1701.23 22560.80 26551.57 24040.85 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
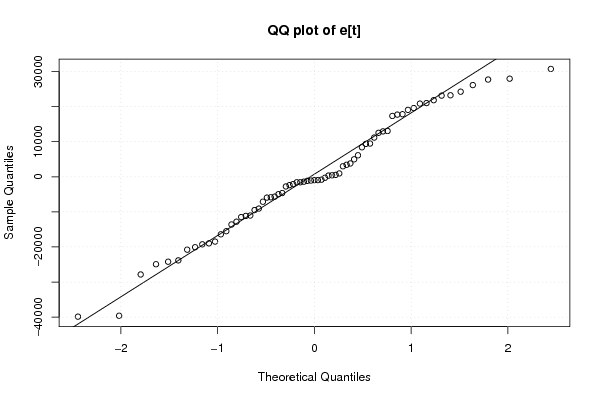
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = 36 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = 36 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||