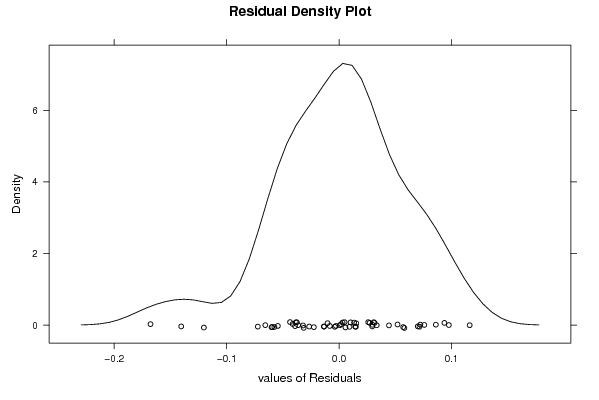
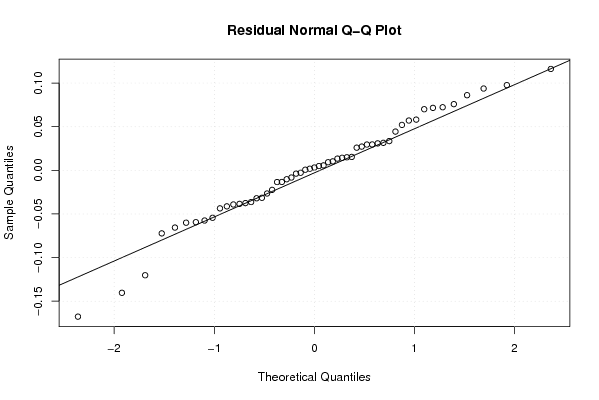
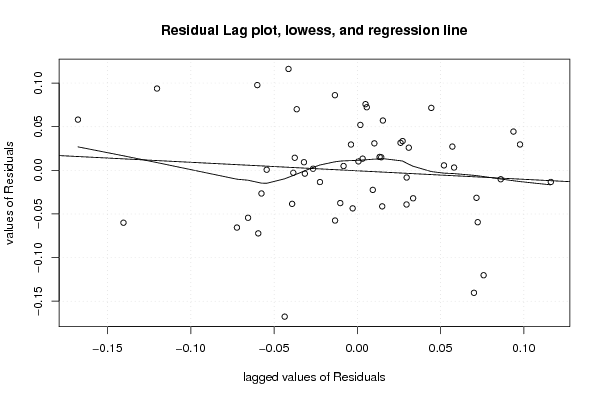
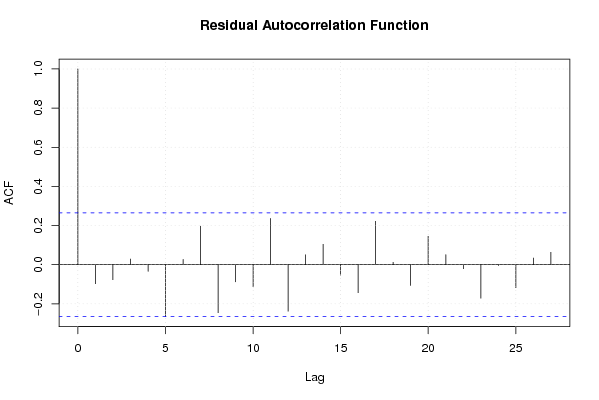
103,91 100,30 103,88 103,77 103,66 103,64 103,63
103,91 98,50 103,91 103,88 103,77 103,66 103,64
103,92 95,10 103,91 103,91 103,88 103,77 103,66
104,05 93,10 103,92 103,91 103,91 103,88 103,77
104,23 92,20 104,05 103,92 103,91 103,91 103,88
104,30 89,00 104,23 104,05 103,92 103,91 103,91
104,31 86,40 104,30 104,23 104,05 103,92 103,91
104,31 84,50 104,31 104,30 104,23 104,05 103,92
104,34 82,70 104,31 104,31 104,30 104,23 104,05
104,55 80,80 104,34 104,31 104,31 104,30 104,23
104,65 81,80 104,55 104,34 104,31 104,31 104,30
104,73 81,80 104,65 104,55 104,34 104,31 104,31
104,75 82,90 104,73 104,65 104,55 104,34 104,31
104,75 83,80 104,75 104,73 104,65 104,55 104,34
104,76 86,20 104,75 104,75 104,73 104,65 104,55
104,94 86,10 104,76 104,75 104,75 104,73 104,65
105,29 86,20 104,94 104,76 104,75 104,75 104,73
105,38 88,80 105,29 104,94 104,76 104,75 104,75
105,43 89,60 105,38 105,29 104,94 104,76 104,75
105,43 87,80 105,43 105,38 105,29 104,94 104,76
105,42 88,30 105,43 105,43 105,38 105,29 104,94
105,52 88,60 105,42 105,43 105,43 105,38 105,29
105,69 91,00 105,52 105,42 105,43 105,43 105,38
105,72 91,50 105,69 105,52 105,42 105,43 105,43
105,74 95,40 105,72 105,69 105,52 105,42 105,43
105,74 98,70 105,74 105,72 105,69 105,52 105,42
105,74 99,90 105,74 105,74 105,72 105,69 105,52
105,95 98,60 105,74 105,74 105,74 105,72 105,69
106,17 100,30 105,95 105,74 105,74 105,74 105,72
106,34 100,20 106,17 105,95 105,74 105,74 105,74
106,37 100,40 106,34 106,17 105,95 105,74 105,74
106,37 101,40 106,37 106,34 106,17 105,95 105,74
106,36 103,00 106,37 106,37 106,34 106,17 105,95
106,44 109,10 106,36 106,37 106,37 106,34 106,17
106,29 111,40 106,44 106,36 106,37 106,37 106,34
106,23 114,10 106,29 106,44 106,36 106,37 106,37
106,23 121,80 106,23 106,29 106,44 106,36 106,37
106,23 127,60 106,23 106,23 106,29 106,44 106,36
106,23 129,90 106,23 106,23 106,23 106,29 106,44
106,34 128,00 106,23 106,23 106,23 106,23 106,29
106,44 123,50 106,34 106,23 106,23 106,23 106,23
106,44 124,00 106,44 106,34 106,23 106,23 106,23
106,48 127,40 106,44 106,44 106,34 106,23 106,23
106,50 127,60 106,48 106,44 106,44 106,34 106,23
106,57 128,40 106,50 106,48 106,44 106,44 106,34
106,40 131,40 106,57 106,50 106,48 106,44 106,44
106,37 135,10 106,40 106,57 106,50 106,48 106,44
106,25 134,00 106,37 106,40 106,57 106,50 106,48
106,21 144,50 106,25 106,37 106,40 106,57 106,50
106,21 147,30 106,21 106,25 106,37 106,40 106,57
106,24 150,90 106,21 106,21 106,25 106,37 106,40
106,19 148,70 106,24 106,21 106,21 106,25 106,37
106,08 141,40 106,19 106,24 106,21 106,21 106,25
106,13 138,90 106,08 106,19 106,24 106,21 106,21
106,09 139,80 106,13 106,08 106,19 106,24 106,21
|