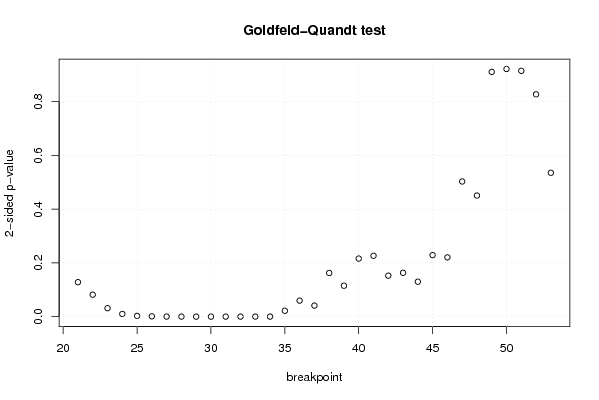
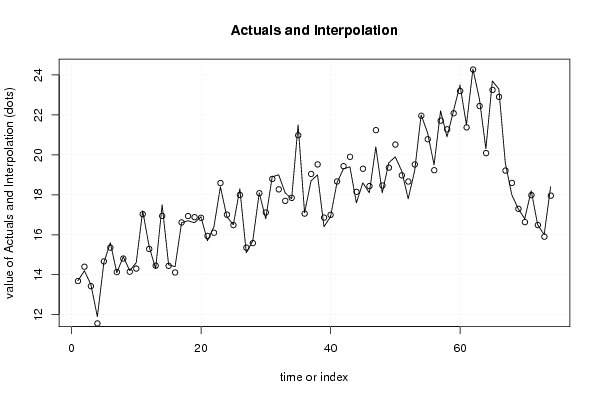
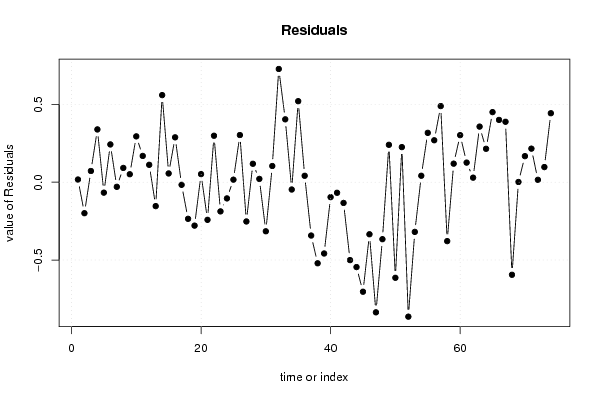
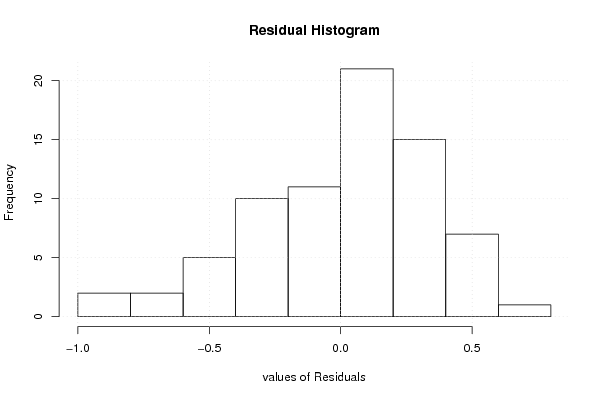
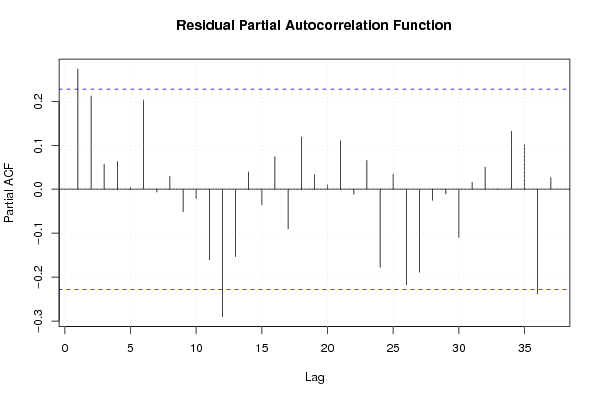
13,7 15 14,4 15,3 14,3 14,5
14,2 15,5 13,7 14,4 15,3 14,3
13,5 15,1 14,2 13,7 14,4 15,3
11,9 11,7 13,5 14,2 13,7 14,4
14,6 16,3 11,9 13,5 14,2 13,7
15,6 16,7 14,6 11,9 13,5 14,2
14,1 15 15,6 14,6 11,9 13,5
14,9 14,9 14,1 15,6 14,6 11,9
14,2 14,6 14,9 14,1 15,6 14,6
14,6 15,3 14,2 14,9 14,1 15,6
17,2 17,9 14,6 14,2 14,9 14,1
15,4 16,4 17,2 14,6 14,2 14,9
14,3 15,4 15,4 17,2 14,6 14,2
17,5 17,9 14,3 15,4 17,2 14,6
14,5 15,9 17,5 14,3 15,4 17,2
14,4 13,9 14,5 17,5 14,3 15,4
16,6 17,8 14,4 14,5 17,5 14,3
16,7 17,9 16,6 14,4 14,5 17,5
16,6 17,4 16,7 16,6 14,4 14,5
16,9 16,7 16,6 16,7 16,6 14,4
15,7 16 16,9 16,6 16,7 16,6
16,4 16,6 15,7 16,9 16,6 16,7
18,4 19,1 16,4 15,7 16,9 16,6
16,9 17,8 18,4 16,4 15,7 16,9
16,5 17,2 16,9 18,4 16,4 15,7
18,3 18,6 16,5 16,9 18,4 16,4
15,1 16,3 18,3 16,5 16,9 18,4
15,7 15,1 15,1 18,3 16,5 16,9
18,1 19,2 15,7 15,1 18,3 16,5
16,8 17,7 18,1 15,7 15,1 18,3
18,9 19,1 16,8 18,1 15,7 15,1
19 18 18,9 16,8 18,1 15,7
18,1 17,5 19 18,9 16,8 18,1
17,8 17,8 18,1 19 18,9 16,8
21,5 21,1 17,8 18,1 19 18,9
17,1 17,2 21,5 17,8 18,1 19
18,7 19,4 17,1 21,5 17,8 18,1
19 19,8 18,7 17,1 21,5 17,8
16,4 17,6 19 18,7 17,1 21,5
16,9 16,2 16,4 19 18,7 17,1
18,6 19,5 16,9 16,4 19 18,7
19,3 19,9 18,6 16,9 16,4 19
19,4 20 19,3 18,6 16,9 16,4
17,6 17,3 19,4 19,3 18,6 16,9
18,6 18,9 17,6 19,4 19,3 18,6
18,1 18,6 18,6 17,6 19,4 19,3
20,4 21,4 18,1 18,6 17,6 19,4
18,1 18,6 20,4 18,1 18,6 17,6
19,6 19,8 18,1 20,4 18,1 18,6
19,9 20,8 19,6 18,1 20,4 18,1
19,2 19,6 19,9 19,6 18,1 20,4
17,8 17,7 19,2 19,9 19,6 18,1
19,2 19,8 17,8 19,2 19,9 19,6
22 22,2 19,2 17,8 19,2 19,9
21,1 20,7 22 19,2 17,8 19,2
19,5 17,9 21,1 22 19,2 17,8
22,2 20,9 19,5 21,1 22 19,2
20,9 21,2 22,2 19,5 21,1 22
22,2 21,4 20,9 22,2 19,5 21,1
23,5 23 22,2 20,9 22,2 19,5
21,5 21,3 23,5 22,2 20,9 22,2
24,3 23,9 21,5 23,5 22,2 20,9
22,8 22,4 24,3 21,5 23,5 22,2
20,3 18,3 22,8 24,3 21,5 23,5
23,7 22,8 20,3 22,8 24,3 21,5
23,3 22,3 23,7 20,3 22,8 24,3
19,6 17,8 23,3 23,7 20,3 22,8
18 16,4 19,6 23,3 23,7 20,3
17,3 16 18 19,6 23,3 23,7
16,8 16,4 17,3 18 19,6 23,3
18,2 17,7 16,8 17,3 18 19,6
16,5 16,6 18,2 16,8 17,3 18
16 16,2 16,5 18,2 16,8 17,3
18,4 18,3 16 16,5 18,2 16,8
|