Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_linear_regression.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Linear Regression Graphical Model Validation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Tue, 27 Oct 2009 14:24:52 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2009/Oct/27/t125667512760r24qfsoubqz8j.htm/, Retrieved Tue, 07 May 2024 13:22:53 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=51218, Retrieved Tue, 07 May 2024 13:22:53 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 132 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Linear Regression Graphical Model Validation] [Workshop 4:Linear...] [2009-10-27 19:42:59] [1433a524809eda02c3198b3ae6eebb69] - D [Linear Regression Graphical Model Validation] [Workshop 4:Linear...] [2009-10-27 20:24:52] [a5c6be3c0aa55fdb2a703a08e16947ef] [Current] - D [Linear Regression Graphical Model Validation] [Workshop 4 part 2...] [2009-10-28 16:52:46] [b6394cb5c2dcec6d17418d3cdf42d699] - D [Linear Regression Graphical Model Validation] [Workshop 4, Part ...] [2009-10-28 16:53:13] [aba88da643e3763d32ff92bd8f92a385] F D [Linear Regression Graphical Model Validation] [workshop 4 part 2] [2009-10-28 16:52:59] [af8eb90b4bf1bcfcc4325c143dbee260] - D [Linear Regression Graphical Model Validation] [Workshop 4 part 2...] [2009-10-28 17:03:12] [b6394cb5c2dcec6d17418d3cdf42d699] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-0.5348 -0.5591 -0.5200 -0.5173 -0.5153 -0.5048 -0.4950 -0.4597 -0.4681 -0.4909 -0.4579 -0.4758 -0.4662 -0.4821 -0.4767 -0.4740 -0.5295 -0.5075 -0.5222 -0.5377 -0.5108 -0.5197 -0.5292 -0.5024 -0.5175 -0.5088 -0.5180 -0.5177 -0.5027 -0.5002 -0.4882 -0.4603 -0.4687 -0.5092 -0.4729 -0.4488 -0.5085 -0.5142 -0.5130 -0.4721 -0.5425 -0.5123 -0.4845 -0.4950 -0.5024 -0.5349 -0.5861 -0.6495 -0.6751 -0.7095 -0.6941 -0.6794 -0.6559 -0.6051 -0.5713 -0.5532 -0.5341 -0.5315 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries Y: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
2.55 2.27 2.26 2.57 3.07 2.76 2.51 2.87 3.14 3.11 3.16 2.47 2.57 2.89 2.63 2.38 1.69 1.96 2.19 1.87 1.60 1.63 1.22 1.21 1.49 1.64 1.66 1.77 1.82 1.78 1.28 1.29 1.37 1.12 1.51 2.24 2.94 3.09 3.46 3.64 4.39 4.15 5.21 5.80 5.91 5.39 5.46 4.72 3.14 2.63 2.32 1.93 0.62 0.60 -0.37 -1.10 -1.68 -0.78 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
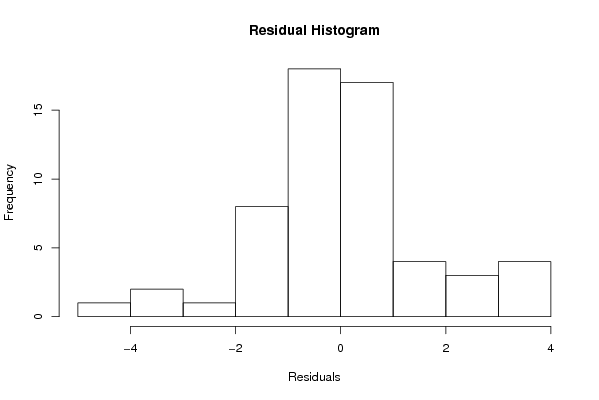
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||