Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *Unverified author* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_density.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
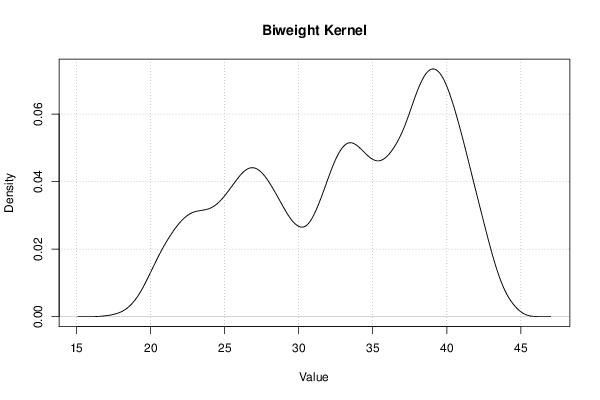
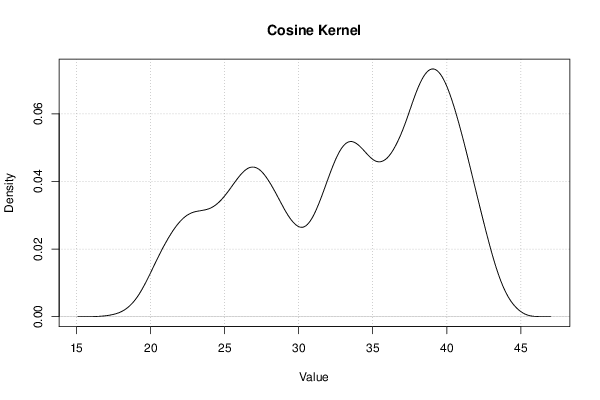
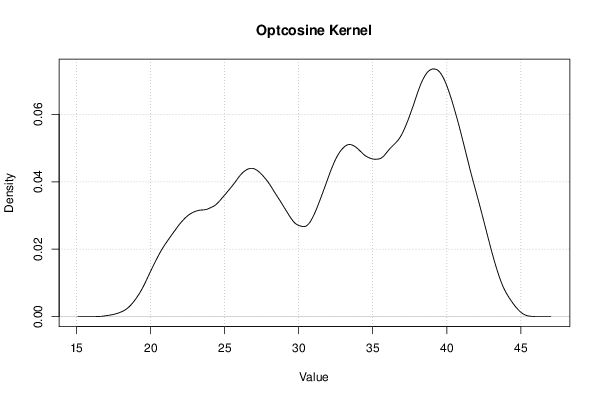
| Title produced by software | Kernel Density Estimation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Fri, 26 Sep 2014 18:23:42 +0100 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2014/Sep/26/t1411752280rnpgvcl9evm2dvv.htm/, Retrieved Sun, 12 May 2024 20:57:45 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=236391, Retrieved Sun, 12 May 2024 20:57:45 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 80 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Kernel Density Estimation] [] [2014-09-26 17:23:42] [1dcac4846cc473f11733ed24aca460fa] [Current] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
42.06 41.85 42 41.85 41.98 41.75 41.77 41.29 41.45 41.9 41.68 41.9 41.77 41.67 41.86 41.8 41.69 41.52 41.6 41.51 41.53 41.41 41.32 41.29 41.29 41.15 41.35 40.79 40.85 39.93 39.7 39.56 39.52 39.34 39.96 39.13 39.34 39.41 39.13 39.53 40.45 40.75 40.99 40.92 40.97 40.98 41.1 42.12 42.14 41.93 42.26 42.41 41.99 42.12 41.56 41.93 41.97 41.96 42.26 42.31 42.39 41.98 41.99 41.91 41.6 41.52 41.46 41.87 41.54 40.79 41.18 40.19 40.39 40.41 40.98 40.84 40.85 40.94 40.73 40.91 40.72 40.75 40.54 40.68 40.7 40.56 40.63 40.77 40.55 40.8 40.6 40.52 40.62 41.02 40.83 40.88 40.78 40.85 40.7 40.52 40.65 40.79 40.79 40.69 40.97 40.82 40.46 40.7 40.7 40.58 40.47 40.43 40.12 39.83 38.71 38.83 38.98 38.84 38.67 38.27 38.24 38.39 38.49 38.53 38.99 38.89 38.51 38.69 38.49 38.48 38.78 38.11 38.36 38.27 38.17 37.95 38.45 38.27 38.84 38.55 38.56 38.4 38.42 38.34 37.88 38.08 37.84 37.9 37.63 37.3 37.59 37.06 37.5 37.98 38.6 38.13 38.85 38.67 38.07 38.17 38.76 37.49 37.44 38.02 37.95 38 38.57 38.77 38.82 39.08 39.61 39.99 39.64 39.72 39.78 39.83 39.64 40 39.96 39.95 40.44 40.48 40.48 40.68 41.12 41.13 40.77 40.69 40.22 40.17 40.1 39.92 39.97 39.26 39.12 39.46 39.47 39.99 39.95 40.26 40.4 40.05 40.28 40.2 40.05 40.1 40.27 40.09 40.49 40.48 40.34 40.2 40.05 40.14 40.24 40.16 40.09 39.82 39.91 40.01 39.74 40.08 39.62 39.36 39.71 39.57 39.51 39.88 39.57 39.02 38.84 39.15 39.08 38.86 38.95 38.58 38.06 38.01 38.18 37.56 37.92 37.77 37.38 37.1 37.08 37.25 37.46 37.81 38 38.19 38.57 38.49 38.65 38.72 39.16 39.33 39.64 38.81 38.81 39.03 38.56 38.59 38.68 38.77 38.52 38.38 38.63 37.88 38.27 38.16 38.47 38.06 37.98 38.6 38.61 38.32 38.63 38.85 39.05 39.03 39.3 39.59 39.93 40.04 40.25 40.3 40.31 40.23 40.18 40.03 40.4 40.41 40.42 40.61 40.65 40.81 41.12 40.95 41.08 40.88 40.86 40.55 39.78 41.05 41.03 41 40.92 40.86 40.65 40.72 40.24 40.47 40.5 40.25 40.65 40.08 39.82 39.45 39.63 40.27 41.01 40.7 40.54 40.38 40.31 40.79 40.88 41.5 41.2 40.55 41.4 41.59 39.95 40.65 41.49 42.34 42.6 41.54 41.93 42.28 42.48 42.95 43.05 42.91 42.5 42.18 42.14 42.11 42.51 42.54 42.21 42.29 42.17 41.66 42.15 42.15 42.24 42.18 42.26 42.64 42.78 42.52 42.28 42.48 42.14 41.4 40.85 40.91 41.13 40.78 40.81 40.06 40.21 40.27 40.78 40.43 40.39 40.38 40.45 40.13 40.11 39.98 39.85 39.4 38.92 38.61 38.82 38.82 38.82 39.28 39.2 39.17 39.04 38.8 38.98 38.6 38.49 38.47 38.03 37.95 38.56 37.76 37.62 37.7 37.51 36.94 37.24 37.67 38.11 38.71 38.85 38.42 38.13 37.56 37.48 37.54 37.53 37.4 36.88 37.07 37.18 37.2 37.08 37.74 37.49 37.43 37.21 36.99 36.95 36.84 37.12 37.2 37.19 37.52 37.65 37.45 36.99 35.88 36.22 36.35 36.7 36.72 37 36.69 37.25 37.53 37.79 37.52 37.7 37.89 37.63 37.7 37.44 37.34 37.2 37.31 37.9 37.94 37.68 37.06 37.44 37.58 37.55 37.33 37.23 36.85 36.49 35.9 36.19 36.03 36.29 36.05 36.65 37.28 36.92 36.95 37.58 37.15 37.16 37.12 37 36.6 36.96 37.37 37.93 37.76 38.06 38.18 38.09 38.26 38.2 38.65 38.5 38.54 38.49 38.46 38.32 38.56 38.16 38.17 38.06 37.78 38.21 37.81 38.73 38.39 38.66 38.38 38.16 38.55 37.73 37.79 37.63 37.96 38.38 37.78 37.355 37.52 37.41 37.38 38.05 38.14 38.56 38.07 38.81 39.14 39.41 39.54 39.66 39.4 39.42 39.6 39.31 39.58 39.735 39.855 40.42 40.505 40.095 40.365 40.485 40.35 39.885 39.525 38.85 38.475 38.45 38.275 38.635 38.76 38.82 39.03 38.55 38.315 38.515 38.95 39.265 39.03 39.01 39.51 39.445 39.05 38.565 38.05 37.62 37.495 37.23 37.475 37.895 37.845 38.125 37.98 38.06 37.395 37.305 37.49 37.86 37.08 37.495 36.695 36.665 36.515 36.995 37.48 37.535 37.865 37.81 37.395 37 37.29 37.105 37.625 38.085 38.33 38.28 38.56 38.6 38.605 38.295 38.505 38.485 38.655 38.68 38.37 38.27 38.21 37.98 37.475 37.27 36.96 36.99 36.98 37.145 36.91 36.59 36.11 36.11 36.075 36.215 36.29 36.48 36.64 36.645 36.835 36.915 36.81 36.175 35.98 35.91 35.87 35.7 35.505 35.24 35.015 35.105 35.19 35.135 35.13 34.755 34.74 34.715 34.59 34.375 34.41 34.51 34.705 34.935 34.45 34.475 34.345 34.675 34.545 34.3 34.44 34.535 34.305 34.615 34.235 34.21 33.765 34.18 34.42 34.045 33.92 34.13 34.035 33.94 33.99 33.645 33.955 34.31 33.955 34.08 34 33.77 33.825 33.73 33.87 33.67 34.21 34.185 34.735 34.5 34.735 34.88 34.955 35.075 35 34.8 34.91 34.905 34.615 34.67 34.26 33.94 33.73 33.735 33.455 33.125 33.545 33.63 33.555 33.35 33.275 33.245 33.47 33.73 33.5 33.67 32.585 32.63 32.575 32.73 32.805 33.345 33.605 33.47 33.835 33.89 33.895 33.985 33.805 33.925 34.145 33.83 33.98 34.05 33.71 33.565 34.325 34.26 34.465 33.645 33.955 34.2 33.97 33.745 33.495 33.62 33.91 34.1 33.655 33.625 33.365 33.28 33.255 32.985 32.735 32.42 33.295 34.225 34.455 34.91 35.06 34.005 33.68 33.685 35.425 35.395 35.035 35.69 34.955 34.67 34.48 34.225 35.305 35.33 35.005 33.715 34.86 35.045 34.935 34.89 34.48 33.965 34.8 34.415 33.735 34.005 33.765 34.125 34.23 33.845 33.735 33.38 32 33.12 32.91 33.09 33.135 33.885 33.5 33.7 34.25 34.18 34.325 34.385 34.675 34.56 34.775 34.41 34.625 33.85 33.63 33.925 33.985 34.085 33.985 33.875 34.175 34.41 34.155 34.04 33.51 33.49 33.095 32.76 32.425 32.535 32.965 33.145 33.055 32.685 32.94 32.485 32.705 32.7 32.55 32.84 32.72 32.615 32.76 32.755 32.635 33.325 33.3 33.47 33.435 33.37 33.64 33.83 33.795 34.16 34.205 34 33.865 34.05 34.135 33.68 33.645 33.5 33.43 33.72 33.85 33.93 33.86 33.88 33.78 33.47 33.685 33.32 33.975 33.945 33.945 33.605 33.785 34.165 33.67 33.616 33.49 33.63 33.695 33.715 33.775 33.75 33.61 33.445 32.98 32.86 32.6 32.6 32.435 32.29 31.645 31.84 31.565 31.485 31.215 31.345 31.475 32.17 32.46 32.625 32.765 32.7 32.66 32.695 32.395 32.295 32.14 32.085 31.885 32.1 31.89 31.68 32.3 31.69 31.54 31.455 31.835 31.615 31.58 32.05 31.215 31.335 31.325 31.35 31.575 31.44 31.16 31.425 31.595 31.585 31.61 31.435 31.59 31.81 31.84 31.605 31.685 31.625 31.605 31.67 31.35 31.39 31.81 31.895 32.51 32.94 32.68 32.715 32.695 32.56 32.775 32.76 32.705 32.675 32.73 32.51 32.425 31.95 32.415 32.195 32.435 32.305 32.035 32.195 32.185 32.4 32.325 31.97 31.475 31.775 32.06 31.92 31.965 32.03 31.8 31.44 31.07 31.455 31.445 31.36 31.245 31.28 31.245 31.25 31.21 30.97 30.915 30.9 30.685 30.575 30.64 30.51 30.395 30.645 30.745 30.505 30.185 29.98 29.905 29.98 29.97 29.86 29.67 29.78 29.74 29.835 29.815 29.575 29.465 29.52 29.43 29.5 29.295 29.375 29.16 29.01 29.075 28.905 28.81 28.825 28.7 28.685 28.925 29.24 29.14 29.115 28.865 28.76 28.68 28.71 28.19 27.6 28.1 27.72 27.715 27.71 27.575 27.755 27.46 27.865 28.055 28.01 27.755 27.82 27.765 28.12 28.32 28.185 28.05 28.205 28.26 28.135 27.91 27.355 27.54 27.525 27.455 27.415 27.1 27.005 26.75 25.985 26.27 26.385 26.36 26.28 26.37 26.08 26.18 25.84 25.27 25.295 25.11 25.15 25.19 25.29 25.365 25.895 26.055 26.28 26.23 26.41 26.195 26.25 25.915 25.915 25.97 25.855 25.93 25.78 25.395 25.615 26.07 26.335 25.77 25.56 25.59 25.305 25.415 25.185 25.605 25.42 26.25 26.5 26.85 26.74 26.68 26.975 26.9 26.81 26.925 26.095 26.835 26.715 26.695 26.855 26.81 26.81 26.505 26.83 27 27.23 27.07 27.285 27.61 27.49 27.115 27.38 27.515 27.34 27.325 26.955 26.89 27.15 27.42 27.78 27.68 27.395 27.45 27.4 27.395 27.455 27.61 27.355 27.245 27.07 26.95 26.91 26.9 26.865 26.845 26.89 27.13 27.17 27.36 27.345 27.35 26.65 26.69 26.61 26.545 26.57 27.5 27.705 27.9 27.9 27.575 27.405 27.255 26.96 26.8 27.3 26.845 26.635 26.62 27.275 27.46 27.265 27.255 27.22 27.355 27 27.035 27.245 27.175 27.775 28.08 28.11 28.475 28.52 28.43 28.045 27.575 27.73 28.165 28.175 28.425 28.58 28.79 28.87 28.725 28.705 28.745 28.6 28.575 28.51 28.63 29.13 29.475 29.575 29.555 29.3 28.99 28.725 28.765 28.48 28.8 29.045 28.915 28.795 28.645 28.49 29.185 29.215 28.855 28.345 28.12 28.465 28.32 28.325 28.03 28.055 28.03 27.675 27.295 26.985 26.755 26.71 26.825 26.755 27.03 26.76 26.625 26.62 26.54 26.955 26.79 26.91 26.795 27.57 27.46 27.365 27.46 27.385 27.415 27.335 27.505 27.43 26.905 26.905 26.53 26.7 26.66 26.7 26.515 26.165 26.29 26.47 26.655 26.74 26.84 26.25 26.22 26.04 25.54 25.065 25.265 25.195 24.97 24.65 24.815 24.275 24.36 24.42 24.81 24.59 24.41 24.56 24.9 24.8 24.695 24.16 24.375 24.22 24.025 24.215 24.48 24.71 24.63 24.83 24.705 24.755 24.78 25.015 24.82 24.91 24.71 24.715 24.68 24.705 24.565 25.16 25.415 25.305 25.155 25.175 24.735 24.51 24.19 24.25 24.36 24.275 24.365 24.34 24.385 24.24 24.105 24.07 23.86 23.71 24.06 24.095 24.26 24.865 23.995 23.94 24.095 24.305 24.34 24.06 24.65 24.755 24.615 24.955 24.57 24.67 24.505 24.625 23.505 23.325 23.565 23.5 23.335 23.32 23.405 23.07 22.57 22.415 21.96 22.015 21.48 21.365 21.475 21.66 21.75 21.66 21.3 21.57 21.565 21.185 21.055 21.21 21.475 21.535 21.6 22.08 22.425 22.625 22.685 22.1 22.3 22.475 22.855 22.345 22.42 22.35 22.685 22.65 21.88 22.135 22.25 22.215 22.475 22.2 21.8 21.58 21.16 20.925 20.65 20.67 20.73 20.54 19.79 19.58 19.815 19.74 19.075 19.58 19.625 19.905 20.225 20.48 21.385 21.47 21.36 21.51 21.485 21.555 21.39 21.5 22.1 21.325 20.48 21.005 21.66 21.365 21.11 21.675 21.25 21.105 21.72 21.85 22 21.64 21.15 21.325 21.32 21.7 21.89 21.95 21.285 21.75 21.9 22.23 22.615 22.42 22.11 22.85 22.785 22.7 22.585 22.31 22.43 22.205 22.025 22.365 22.365 22.725 23.035 23.04 22.635 22.44 21.875 22.59 22.235 22.765 23.245 22.165 22.785 21.835 22.51 23.095 22.6 22.13 22.915 22.245 20.82 21 21.825 21.66 22.31 22.795 21.925 21.845 22.795 23.435 22.42 22.375 22.86 23.35 22.255 22.57 22.47 22.3 20.78 20.575 20.5 22.8 22.8 23.18 22.545 22.48 22.4 23.415 23.55 21.715 21.025 23.785 24.325 25.65 25.67 26.89 26.525 26.31 25.905 26.04 25.47 25.2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
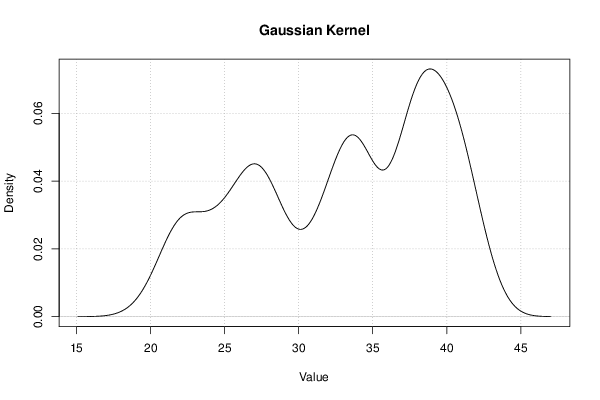
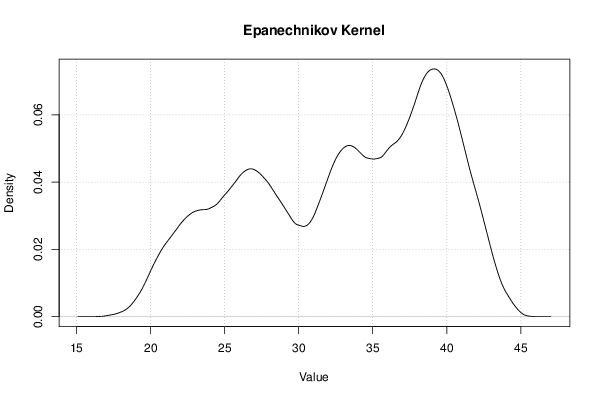
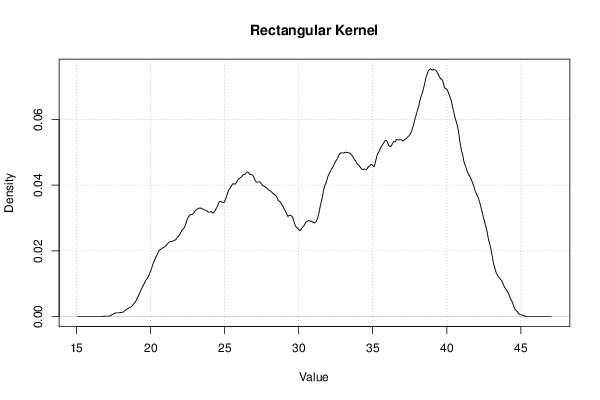
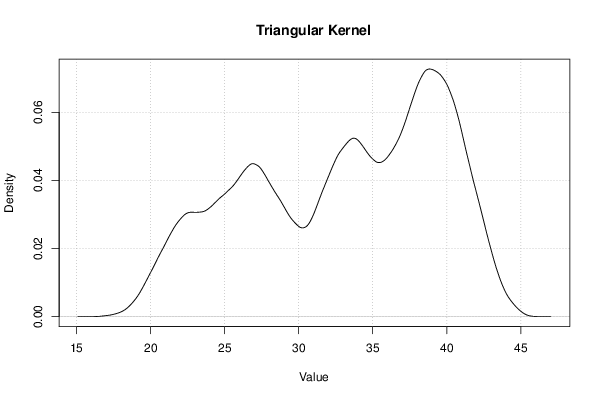
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = no ; par3 = 512 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = no ; par3 = 512 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
if (par1 == '0') bw <- 'nrd0' | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||